Abstract
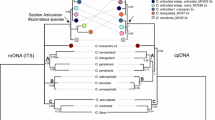
The economically important genus Arachis (Fabaceae) comprises 80 species restricted to South America. One monograph on the genus divided it into nine sections and included an intuitive assessment of evolutionary relationships. There is no comprehensive phylogenetic study of the genus. To test the current systematic treatment of the genus, we reconstructed a phylogeny for Arachis using nuclear ITS and plastid trnT–trnF sequences from 46 species representing all nine sections. ITS cloning of the allotetraploid species of section Arachis indicated the presence of A and B genome alleles and chimeric sequences. Our study revealed that species from section Extranervosae were the first emerging lineage in the genus, followed by sections Triseminatae and Caulorrhizae, and two terminal major lineages, which we refer to as erectoides and arachis. The lineage erectoides comprises members of sections Erectoides, Heteranthae, Procumbentes, Rhizomatosae, and Trierectoides. Species in the arachis lineage form two major clades, arachis I (B and D genomes species and the aneuploids) and arachis II (A genome species). Our results substantiated the sectional treatment of Caulorrhizae, Extranervosae, and Triseminatae, but demonstrated that sections Erectoides, Procumbentes, and Trierectoides are not monophyletic. A detailed study of the genus Arachis with denser taxon sampling, additional genomic regions, plus information from morphology and cytogenetics is needed for comprehensive assessment of its systematics.


Similar content being viewed by others
References
Álverez I, Wendel JF (2003) Ribosomal ITS sequence and plant phylogenetic inference. Mol Phylogenet Evol 29:417–434
Angelici C, Hoshino AA, Nobile PM, Palmieri DA, Valls JFM, Gimenes MA, Lopes CR (2008) Genetic diversity in section Rhizomatosae of the genus Arachis (Fabaceae) based on microsatellite markers. Genet Mol Biol 31:79–88
Baker FK, Lutzoni FM (2002) The utility of the incongruence length difference test. Syst Biol 51:625–637
Barkley NA, Dean RE, Pittman RN, Wang ML, Holbrook CC, Pederson GA (2007) Genetic diversity of cultivated and wild-type peanuts evaluated with M13-tailed SSR markers and sequencing. Genet Res 89:93–106
Bertozo MR, Valls JFM (2001) Seed storage protein electrophoresis in Arachis pintoi and A. repens (Leguminosae) for evaluating genetic diversity. Genet Resour Crop Evol 48:121–130
Birnboim HC, Dolye J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucl Acids Res 7:1513–1523
Borsch T, Quandt D (2009) Mutational dynamics and phylogenetic utility of non-coding plastid DNA. Plant Syst Evol Special Issue on non-coding DNA. Evolution 282:169–199
Bravo JP, Hoshino AA, Angelici C, Lopes CR, Gimenes MA (2006) Transferability and use of microsatellite markers for the genetic analysis of the germplasm of some Arachis section species of the genus Arachis. Genet Mol Biol 29:516–524
Burow M, Simpson C, Faries M, Starr J, Paterson A (2009) Molecular biogeographic study of recently described B- and A-genome Arachis species, also providing new insights into the origins of cultivated peanut. Genome 52:107–119
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Clement M, Snell Q, Walke P, Posada D, Crandall KA (2002) TCS: estimating gene genealogies. In: Parallel distributed processing symposium (Proceedings international IPDPS. Abstracts and CD-Rom), pp 184–190
Creste S, Tsai S, Valls J, Gimenes M, Lopes C (2005) Genetic characterization of Brazilian annual Arachis species from sections Arachis and Heteranthae using RAPD markers. Genet Resour Crop Evol 52:1079–1086
Darlu P, Lecointre G (2002) When does the incongruence length difference test fail? MBE 19:432–437
Dolphin K, Belshaw R, Orme CDL, Quicke DLJ (2000) Noise and incongruence: interpreting results of incongruence length difference test. Mol Phylogenet Evol 17:401–406
Farris JS, Kallersjo M, Kluge AG, Bult C (1995) Testing significance of incongruence. Cladistics 10:315–319
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fernández A, Krapovickas A (1994) Cromosomas y evolucion en Arachis (Leguminosae). Bonplandia 8:187–220
Galgaro L, Valls JFM, Lopes CR (1997) Study of the genetic variability and similarity among and within Arachis villosulicarpa, A. pietrarellii and A. hypogaea through isoenzyme analysis. Genet Resour Crop Evol 44:9–15
Galgaro L, Lopes CR, Gimenes M, Valls JFM, Kochert G (1998) Genetic variation between several species of section Extranervosae, Caulorrhizae, Heteranthae, and Triseminatae (genus Arachis) estimated by DNA polymorphism. Genome 41:445–454
Gimenes MA, Lopes CR, Galgaro L, Valls JFM, Kochert G (2002a) RFLP analysis of genetic variation in species of section Arachis, genus Arachis (Leguminosae). Euphytica 123:421–429
Gimenes MA, Lopes CR, Valls JFM (2002b) Genetic relationships among Arachis species based on AFLP. Genet Mol Biol 25:349–353
Gimenes MA, Hoshino AA, Barbosa AV, Palmieri DA, Lopes CR (2007) Characterization and transferability of microsatellite markers of the cultivated peanut (Arachis hypogaea). BMC Pl Biol 7:9
Gregory WC, Gregory MP (1979) Exotic germ plasm of Arachis L. interspecific hybrids. J Hered 70:185–193
Gregory WC, Gregory MP, Krapovicaks A, Smith BW, Yarbrough JA (1973) Structure and genetic resources of peanuts. In: Peanuts—culture and uses. American Peanut Research and Education Association, Inc., Stillwater, pp 47–133
Gregory WC, Krapovicaks A, Gregory MP (1980) Structure, variation, evolution and classification in Arachis. In: Summerfield RJ, Bunting AH (eds) Advances in legume sciences. Royal Botanical Gardens, Kew, pp 469–481
Halward TM, Stalker HT, LaRue EA, Kochert G (1991) Genetic variation detectable with molecular markers among unadapted germ-plasm resources of cultivated peanut and related wild species. Genome 34:1013–1020
Hammer K, Arrowsmith N, Gladis T (2003) Agrobiodiversity with emphasis on plant genomics research. Naturwissenschaften 90:241–250
Hilu KW, Stalker HT (1995) Genetic relationships between peanut and wild species of Arachis sect. Arachis (Fabaceae): evidence from RAPDs. Pl Syst Evol 198:167–178
Hoshino AA, Bravo JP, Angelici CM, Barbosa AVG, Lopes CR, Gimenes MA (2006) Heterologous microsatellite primer pairs informative for the whole genus Arachis. Genet Mol Biol 29:665–675
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Johnson LA, Soltis DE (1998) Assessing congruence: empirical examples from molecular data. In: Soltis DE, Soltis PS, Doyle JJ (eds) Molecular systematics of plants II: DNA sequencing. Kluwer, Boston, pp 297–348
Jung S, Tate PL, Horn R, Kochert G, Moore K, Abbott AG (2003) The phylogenetic relationship of possible progenitors of the cultivated peanut. J Hered 94:334–340
Kelchner SA (2000) The evolution of non-coding chloroplast DNA and its application in plant systematics. Ann Mo Bot Gard 87:482–498
Kochert G, Halward T, Branch WD, Simpson CE (1991) RFLP variability in peanut (Arachis hypogaea L) cultivars and wild species. Theor Appl Genet 81:565–570
Kovarik A, Matyasek R, Lim K, Skalicka K, Koukalova B, Knapp S, Chase M, Leitch A (2004) Concerted evolution of 18-5.8-26S rDNA repeats in Nicotiana allotetraploids. Biol J Linn Soc 82:615–625
Krapovickas A, Gregory WC (1994) Taxonomía del género Arachis (Leguminosae). Bonplandia 8:1–186
Lavia GI (1998) Karyotypes of Arachis palustris and A. praecox (Section Arachis), two species with basic chromosome number x = 9. Cytologia 63:177–181
Lavin M, Pennington RT, Klitgaard BB, Sprent JI, de Lima HC, Gasson PE (2001) The dalbergioid legumes (Fabaceae): delimitation of a pantropical monophyletic clade. Am J Bot 88:503–533
Milla SR, Isleib TG, Stalker HT (2005) Taxonomic relationships among Arachis sect. Arachis species as revealed by AFLP markers. Genome 48:1–11
Moretzsohn MDC, Hopkins MS, Mitchell SE, Kretovich S, Valls JFM, Ferreira EM (2004) Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on the analysis of hypervariable regions of the genome. BMC Pl Biol 4:11
Müller K (2005) SeqState: primer design and sequence statistics for phylogenetic DNA data sets. Appl Bioinf 4:65–69
Müller K (2007) PRAP2—likelihood and parsimony rachet analysis, v.09
Müller J, Müller K (2004) TreeGraph: automated drawing of complex tree figures using an extensible tree description format. Mol Ecol Notes 4:786–788
Müller K, Quandt D, Müller J, Neinhuis C (2005) PhyDE, version 0.92: phylogenetic data editor
Nixon K (1999) The parsimony ratchet, a new method for rapid parsimony analysis. Cladistics 15:407–414
Nóbile PM, Gimenes MA, Valls JFM, Lopes CR (2004) Genetic variation within and among species of genus Arachis, section Rhizomatosae. Genet Resour Crop Evol 51:299–307
Palmieri DA, Bechara MD, Curi RA, Gimenes MA, Lopes CR (2005) Novel polymorphic microsatellite markers in section Caulorrhizae (Arachis, Favaceae). Mol Ecol Notes 5:77–79
Peñaloza APS, Valls JFM (1997) Contagen do número cromossômico en assos de Arachis decora (Legumonsae). In: Vega RFA, Bovi MLA, Betti JA, Voltan RBQ (eds) Simpósio Latino Slericanno de Recursos Genéticos Vegetais. IAC/Embrapa-Cenargen, Campanias, p 21
Peñaloza APS, Valls JFM (2005) Chromosome number and satellited chromosome morphology of eleven species of Arachis (Leguminosae). Bonpladia 15:65–72
Quandt D, Müller K, Huttenen S (2003) Characterisation of the chloroplast DNA psbT-H region and the influence of dvad symmestrical elements on phylogentic reconstructions. Pl Biol 5:400–410
Quandt D, Müller K, Stech M, Frahm J-P, Frey W, Hilu KW, Borsch T (2004) Molecular evolution of the chloroplast trnL–F region in land plants. Monogr Syst Bot Mo Bot Gard 98:13–37
Raina S, Mukai Y (1999) Detection of a variable number of 18S-5.8 S-26S and 5S ribosomal DNA loci by fluorescent in situ hybridization in diploid and tetraploid Arachis species. Genome 42:52–59
Seijo JG, Lavia GI, Fernandez A, Krapovickas A, Ducasse D, Moscone EA (2004) Physical mapping of the 5S and 18S-25S rRNA genes by FISH as evidence that Arachis duranensis and A. ipaensis are the wild diploid progenitors of A. hypogaea (Leguminosae). Am J Bot 91:1294–1303
Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analysis. Syst Biol 49:369–381
Singh AK, Simpson CE (1994) Biosystematics and genetic resources. In: Smartt J (ed) The groundnut crop: a scientific basis for improvement. Chapman & Hall, London, pp 96–137
Smartt J, Gregory WC, Gregory MP (1978) The genomes of Arachis hypogaea L. 1. Cytogenetic studies of putative genome donors. Euphytica 27:665–675
Soltis DE, Mavrodiev EV, Doyle JJ, Rauscher J, Soltis PS (2008) ITS and ETS sequence data and phylogeny reconstruction in allopolyploids and hybrids. Syst Bot 33:7–20
Stalker HT (1981) Hybrids in the genus Arachis between sections Erectoides and Arachis. Crop Sci 21:359–362
Stalker HT (1991) A new species in section Arachis of peanuts with a D genome. Am J Bot 78:630–637
Stalker HT, Moss JP (1987) Speciation, cytogenetics, and utilization of Arachis species. Adv Agron 41:1–40
Stöver B, Müller K (2010) TreeGraph2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinf 11:7
Swofford DL (2003) PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4.0, β. Sinauer Associates, Sunderland
Taberlet P, Gilley L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of the chloroplast DNA. Pl Mol Biol 17:1105–1109
Tallury SP, Hilu KW, Milla SR, Friend SA, Alsaghir M, Stalker HT, Quandt D (2005) Genomic affinities in Arachis section Arachis (Fabaceae): molecular and cytogenetic evidence. Theor Appl Genet 111:1229–1237
Tun YT, Yamaguchi H (2007) Phylogenetic relationship of wild and cultivated Vigna (Subgenus Ceratotropis, Fabaceae) from Myanmar based on sequence variations in non-coding regions of trnT–F. Breed Sci 57:271–280
Valls JFM, Simpson CE (1994) Taxonomy, natural distribution and attributes of Arachis. In: Kerridge PC, Hardy B (eds) Biology and agronomy of forage Arachis. CIAT, California, pp 1–18
Valls JFM, Simpson CE (2005) New species of Arachis (Leguminosae) from Brazil, Paraguay and Bolivia. Bonplandia 14:35–63
Vander Stappen J, Van Campenhout S, Gama Lopez S, Volckaert G (1998) Sequencing of the internal transcribed spacer region ITS1 as a molecular tool detecting variation in the Stylosanthes guianensis species complex. Theor Appl Genet 96:869–877
Vander Stappen J, De Laet J, Gama-López S, Van Campenhout S, Volckaert G (2002) Phylogenetic analysis of Stylosanthes (Fabaceae) based on the internal transcribed spacer region (ITS) of nuclear ribosomal DNA. Pl Syst Evol 234:27–51
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Wojciechowski MF, Lavin M, Sanderson MJ (2004) A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot 91:1846–1862
Acknowledgements
We thank the USDA Southern Regional PI station for some of the plant material used for this study, Deborah Wiley for growing and maintaining plant samples, and Chelsea M. Black and Daniel S. Volpe for assisting in laboratory work. This work was partially funded by the Virginia Academy of Science, the American Society of Plant Taxonomists, and the Virginia Tech Graduate Research Development Program.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Friend, S.A., Quandt, D., Tallury, S.P. et al. Species, genomes, and section relationships in the genus Arachis (Fabaceae): a molecular phylogeny. Plant Syst Evol 290, 185–199 (2010). https://doi.org/10.1007/s00606-010-0360-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-010-0360-8