Microbes: A Hidden Treasure of Polyunsaturated Fatty Acids
- 1Colin Ratledge Center of Microbial Lipids, School of Agriculture Engineering and Food Sciences, Shandong University of Technology, Zibo, China
- 2Department of Botany and Microbiology, Faculty of Science, Al-Azhar University, Assiut, Egypt
- 3Institute of Agricultural Facilities and Equipment, Jiangsu Academy of Agricultural Sciences, Nanjing, China
Microbes have gained a lot of attention for their potential in producing polyunsaturated fatty acids (PUFAs). PUFAs are gaining scientific interest due to their important health-promoting effects on higher organisms including humans. The current sources of PUFAs (animal and plant) have associated limitations that have led to increased interest in microbial PUFAs as most reliable alternative source. The focus is on increasing the product value of existing oleaginous microbes or discovering new microbes by implementing new biotechnological strategies in order to compete with other sources. The multidisciplinary approaches, including metabolic engineering, high-throughput screening, tapping new microbial sources, genome-mining as well as co-culturing and elicitation for the production of PUFAs, have been considered and discussed in this review. The usage of agro-industrial wastes as alternative low-cost substrates in fermentation for high-value single-cell oil production has also been discussed. Multidisciplinary approaches combined with new technologies may help to uncover new microbial PUFA sources that may have nutraceutical and biotechnological importance.
Introduction
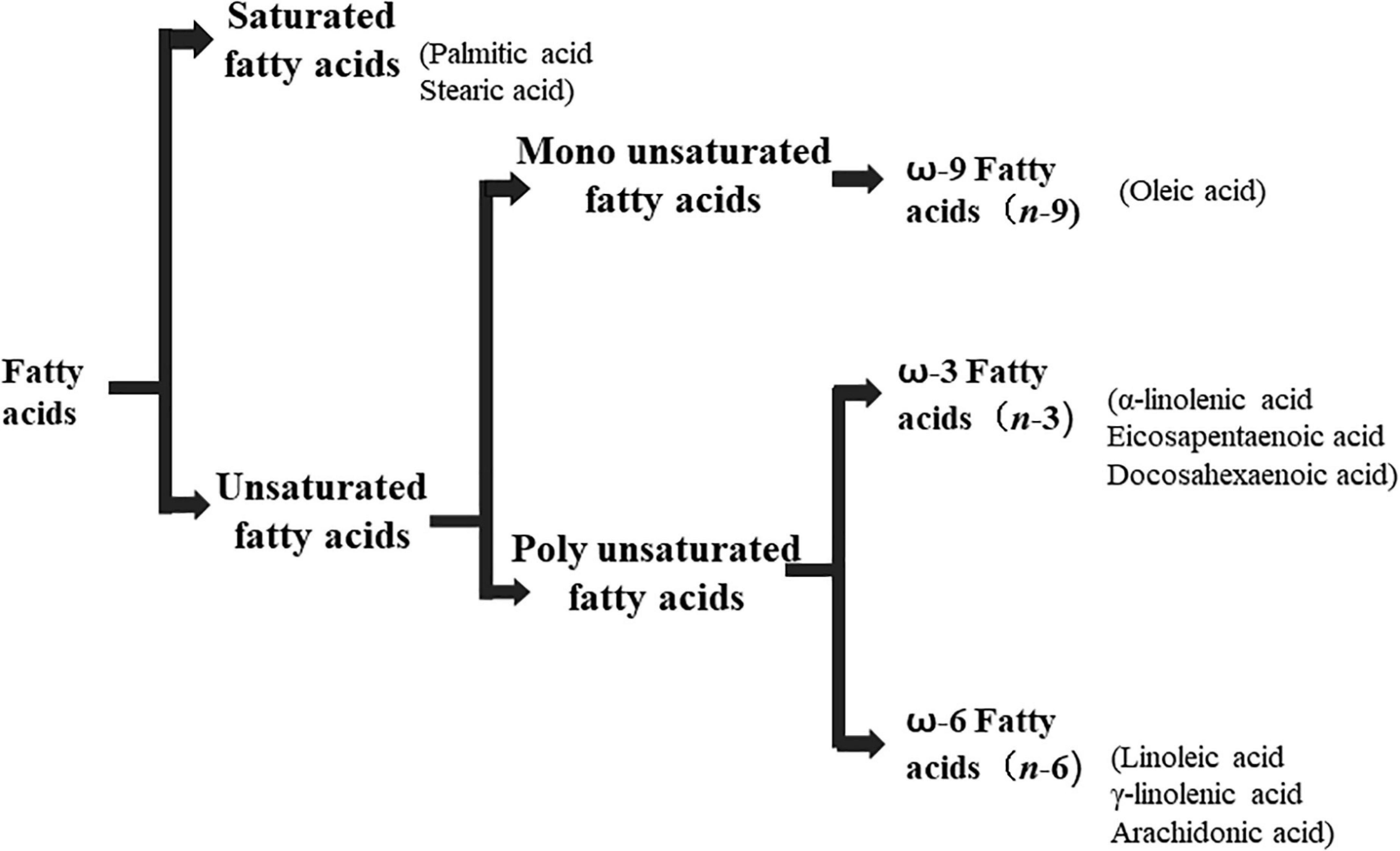
Lipids include oils, fats, phospholipids, steroids, waxes, and certain related compounds are characterized by water insolubility and nonpolar organic solvents solubility (1). Lipids have a biological significance for their involvement in cell membranes, energy source, signaling pathways, hormones, vitamins, gene expression, and bioactive lipid mediator synthesis (2). Lipids are an important component of the daily diet as they provide energy as well as needed fatty acids (3). Fatty acids (FAs) are the simplest lipids, organic compounds containing hydrophobic hydrocarbon chain varying in carbon atoms from C6 to C32 attached to a hydrophilic carboxyl functional group. The majority of FAs seen in nature contain linear hydrocarbon chains, while a few, mainly in bacteria, may have branching or even cyclic structures. FAs on the basis of presence or absence, number of their double bonds can be categorized into saturated fatty acids or unsaturated fatty acids (Figure 1). Dietary saturated FAs and MUFAs are mainly supplied from foods of plant and animal origin. Trans-fatty acids containing a double bond in a trans-configuration are the most common type of MUFA derived via industrial hydrogenation (4). In light of therapeutic and physiological properties, dietary polyunsaturated fatty acids (PUFAs) are regarded as being extremely significant (5). The two significant ω-6 PUFAs are arachidonic acid and linoleic acid constitute 85%, whilst the ω-3 PUFAs, such as docosahexaenoic acid, eicosapentaenoic acid, and alpha-linolenic acid constitute the remainder of the 10–15% total PUFAs (6). The human body cannot synthesize most PUFAs and they are therefore called as essential fatty acids and must be supplied through diet. On the other hand, saturated and MUFAs can be synthesized by the human body, therefore considered non-essential fatty acids (7). Fatty acid production can occur via aerobic metabolic pathways found in fungi, algae, plants, and animals, or anaerobic pathways using polyketide synthase enzymes found in some bacteria and algae. This involves both elongase and desaturase enzyme-controlled elongation and desaturation processes (8).
Polyunsaturated fatty acids play a vital role in human health, including nervous system development, cardioprotective functions, inflammatory disorders, assisting in the reduction of serum triglyceride concentrations, and cancer prevention (9). PUFAs are found in all animals and many plants, microorganisms, particularly algae, fungi, and bacteria, have the highest diversity and offers excellent alternatives for the production of nutritionally important PUFAs (5, 10). PUFAs from oleaginous microbes can be economically feasible and sustainable than animals and plants and are the target of most industrial microbial engineering (11).
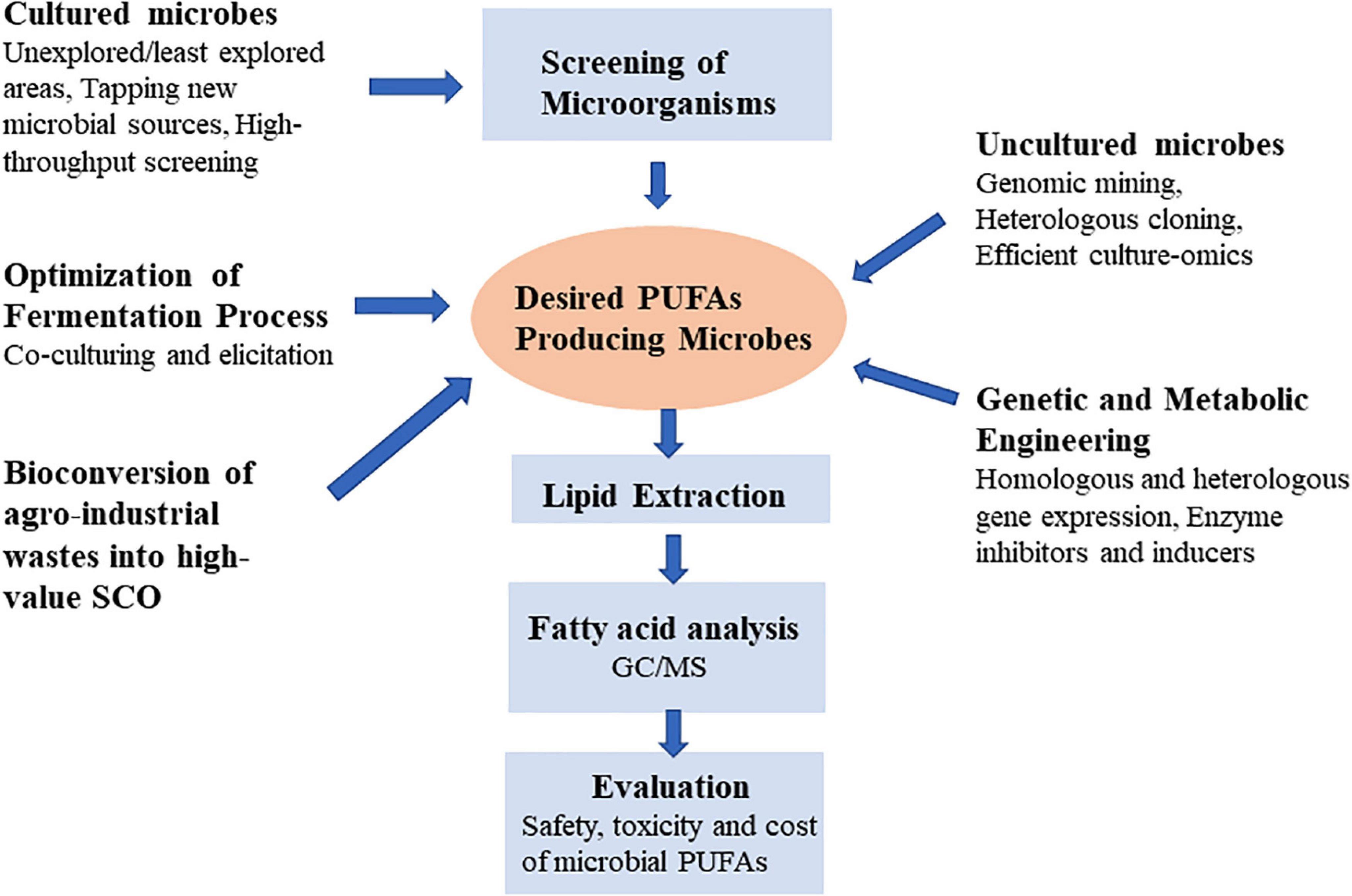
Lipid biotechnology is emerging as a significant subject of research due to advances in analytical techniques and genetic engineering, as well as diverse application of lipids and their derivatives in different industries. Modern lipidomics techniques, such as GC, LC, EI, MS, NMR, and MALDI can rapidly screen a large number of microbes for their lipid composition (8, 12, 13). In microbial biotechnology, the biggest problems of PUFAs production are to improve yields to meet cost competitiveness demands of different sectors (14). New perspectives for the development of sustainable microorganisms producing PUFAs would be opened up by better understanding of microbiology, analytical techniques and metabolic engineering. The benefits of study on the PUFAs in oleaginous microbes might be overwhelming from fundamental research to industrial applications. This review outlines the fundamental features of the generation of PUFA microbial cell factory. It addresses the relevance, importance, genetic modification, and novel strategies for achieving more PUFA production in microorganisms. Additionally, it also discusses how we can use more effective methodologies to find new PUFA-producing microbial strains by mining less explored natural resources (Figure 2).
Importance of Polyunsaturated Fatty Acids
Polyunsaturated fatty acids are fatty acids with more than one double bond in their chemical structure and are categorized into n-3 (ω-3) and n-6 (ω-6) depending on where the first double bond is located on the methyl end of the FA chain. PUFAs are more commonly found in cis-form in nature. Unsaturated fatty acids in membranes are often restricted to the phospholipids’ sn-2 (sn = stereospecific numbering) position with 18 to 20 carbon atoms. PUFAs are key constituent of cell membranes, glycolipids and phospholipids, precursors for many regulatory molecules and hormones, and have been recognized as health-promoting active components as shown in Table 1 (5, 15–17).
According to several medical studies, PUFAs are essential for brain, nerve, eye function (18, 19), prevention of neurodegenerative diseases and psychiatric disorders (20), as well as for the prevention of cardiovascular diseases (21), infection (22), and cancer (9). The American Heart Association (AHA) recommends eating oily fish twice a week for adults. Fish, particularly oily species have high levels of EPA and DHA that have been found to be cardioprotective. EPA has the main human benefits of reducing the risk of colon, breast and pancreatic cancers, lowering plasma cholesterol, protects against atherosclerosis, antiaggregatory property, protecting against cardiovascular illnesses, and is vital in maintaining homeostasis (23–26). DHA is essential for appropriate neurological and visual development in babies, as well as protection against cardiovascular disease (23, 27). GLA has also been reported to have anti-cancer properties, particularly against breast cancer and malignant glioma cells (28). Direct transformation studies on microbial lipids have resulted in the production of PUFAs salts that showed potent bioactivities than parent PUFAs. For example the lipids containing either GLA or EPA produced by N. salina and T. elegans were transformed into water-soluble fatty acid potassium salts (FAPS) and evaluated in vitro against a variety of human pathogenic bacteria and the MCF-7 cancer cell line. FAPS produced from the lipids were found to be potent inhibitor of various Gram positive and Gram negative bacteria, as well as breast cancer cells, even at low doses (29). Similarly, fatty acid lithium salts (FALS) derived from C. echinulata lipids containing GLA showed high cytotoxicity and genotoxicity inducing DNA fragmentation of HL-60 cancer cells (30). ARA performs several key physiological functions as a biogenetic precursor to the bio-active prostaglandins, prostacyclins, thromboxans, and leucotrienes (31). The Food and Agriculture Organization/World Health Organization (FAO/WHO) advised that newborn milk be supplemented with arachidonic acid (32). PUFAs also have shown antimicrobial role, inhibiting the proliferation of certain food borne and food spoilage microorganisms (33, 34). PUFAs presence in the diet improves the absorption of vitamins A, D, E, and K (fat-soluble vitamins) and also regulates cholesterol metabolism (28).
Why Microbial Polyunsaturated Fatty Acids?
Due to the difficulty of synthetically inserting cis double bonds into saturated fatty acids, PUFAs are considered a natural product (35). According to current projections, DHA availability may decline to the point where almost 90% population would face a shortage (16). Notably, there is a significant discrepancy between demand and supply, with the deficit anticipated to be more than 1 million tons (36). Researchers throughout the world are exploring alternative sources of PUFAs and microbes have been identified as promising sources (37). Microbial production of PUFAs has attracted more attention and various companies are underway for their commercial production and marketing. In 2002, Photon Company (was founded in New Zealand to commercialize microbial EPA. Microbial PUFAs have an advantage in comparison to plant and fish oil, as microbes uses different substrates including wastes for their growth and can be cultured regardless of climate change. Microorganisms, due to their fast growth and structural simplicity, can be utilized as a model to research the PUFA metabolic pathway, leading to a better knowledge of the process and simple to manipulate in order to maximize productivity. The production via fermentation of microbial PUFAs is an excellent and renewable source. Some bacteria, fungus and marine algae are key providers of PUFAs in the microbial world (38). Numerous species of bacteria, fungi and microalgae can accumulate 20% w/w or more lipids in their cellular compartment; these organisms are known as oleaginous microorganisms (39). The majority of lipids contained in oleaginous bacteria are 4 to 28 saturated or unsaturated fatty acids of non-branched carbon chains (40, 41). Based on their fatty acid compositions, oleaginous microorganisms can be used in the biodiesel or nutraceutical industries. Oleaginous microbes can ferment low-value agro-industrial raw materials into value-added products lipids in either a solid or submerged process (42, 43). All microorganisms have the potential to synthesize lipid, but only a few of them are naturally capable of synthesizing and storing extra lipid, including PUFAs, as shown in Table 2.
Bacteria often produce a single type of PUFA rather than a mixture, requiring less purification and hence lowering production costs. Discovery of bacteria produces EPA and DHA from sediments of deep sea water has drawn scientific interest, which has in turn paved the way for the use of bacterial PUFAs in the nutraceutical industry (74, 75). In bacterial cells, EPA is found in the form of phospholipids such as phosphatidyl ethanolamine, phosphatidyl glycerol, and cardioolipin (76). To date, the majority of PUFA-producing bacteria have been discovered as belonging to Photobacterium, Shewanella, Moritella, Colwellia, Psychromonas, Alteromonas, and Vibrio (75, 77–79). It was discovered that the majority of PUFA manufacturers are topiezophilic and psychrophilic, with origins in the deep sea and polar regions (74). Increased membrane fluidity due to high PUFA content in the plasma membrane is essential for such microbes to adapt to harsh environments (74, 77).
Among fungi order Mucorales have largest number of species that are typically fast-growing, saprotrophic and many of its members were reported oleaginous (80). Zygomycete fungi produce important PUFAs such as GLA, DGLA, ARA, and EPA (81, 82). It is therefore no surprise Mortierella alpina, Yarrowia lipolytica, and Mucor circinelloides are among the best PUFA producers in the world. Even better, the list of producers is always growing. Mucor circinelloides, a filamentous fungus, was the first microbial strain identified to produce commercial GLA rich oil (83). M. alpina, an oleaginous fungus, is a good source of ARA and has been exploited commercially since late 1980s to generate ARA-rich oil for infant formula (84). Y. lipolytica is labeled as safe and is regarded as best host for the manufacturing of PUFAs in nutraceutical industries (84).
When it comes to producing PUFAs, microalgae are excellent source, due to their ability to thrive in the presence of nutrient- and oxygen-controlled conditions as well as carbon dioxide- and light-controlled environments (85). The photoautotrophic marine oleaginous diatom Fistulifera solaris has been shown to accumulate large percentage of EPA in their dry biomass (86). Diatom Nitzschia laevis has been reported to produce large quantities of EPA (87). Phaeodactylum tricornutum, a photoautotrophic diatom, produced more EPA and DHA when cultivated in a mixotrophic environment than when grown in a photoautotrophic environment. Crypthecodinium cohnii, a marine microalgal heterotrophic species that has been utilized for DHA production on commercial scale, is another promising candidate (88). The ability of marine microalgae to accumulate substantial amount triacylglycerides (TAGs) rich in PUFAs is remarkable. Unicellular Thraustochytrids such as Schizochytrium, Thraustochytrium, and Aurantiochytrium are important heterotrophic microalgae that accumulate significant amounts of DHA (89). Photosynthetic microalgae, like Dunaliella salina (90) Nannochloropsis oceanica (91), and Phaeodactylum tricornutum (92) primarily synthesize EPA (93). LA and ALA are primarily stored by other species, such as the green algae Chlorella vulgaris (94).
Metabolic Engineering of Existing Microbes
In microorganisms, two separate metabolic mechanisms, desaturase/elongase or aerobic pathway and polyketide synthase (PKS) or anaerobic pathway, are responsible for the production of PUFAs (95). Several sets of genes encoded enzymes desaturase and elongase isolated and identified from the majority of eukaryotic and prokaryotic species, including animals, plants, and the majority of other microbes, showed aerobic pathway of PUFAs biosynthesis (96, 97). The D4-, D5-, D6-, D8-, D12-, and D15-desaturases have largely been found in animals, plants, and microbes (98–100). The detected elongases are classified into two groups: FAE1-like and ELO-like sequences. FAE1-like sequences are membrane-bound, and the most common plant elongases were shown to be selective for saturated and MUFAs (101). On the other hand, ELO-like sequences like ELO1, ELO2, and ELO3 are three distinct yeast elongases that have varied fatty acid substrate preferences (102). In non-photosynthetic microbes de novo fatty acid synthesis takes pace cytoplasm while as in the plastids of photosynthetic microbes. Enzymes for fatty acid desaturation and elongation are mostly associated with the endoplasmic reticulum membrane in eukaryotes (103).
Instead of using the desaturase/elongase pathway for PUFA biosynthesis, some prokaryotic and eukaryotic microorganisms use PKS to synthesize DHA or EPA referred as an alternative PUFA biosynthetic pathway (95). This route was first reported in Shewanella pneumatophori strain SCRC-2378, an EPA producing marine bacteria, which had PUFA levels ranging from 24 to 40% of total fatty acids (104). The PKS genes are thought to encode multi-functional complexes that can synthesis EPA without the assistance of FAS, aerobic desaturase/elongase. There were eight closely similar PKS proteins and three homologous bacterial FAS proteins in the protein sequences encoded by ORFs within the big DNA fragment (95). Various bacteria inhabiting in the deep sea such as Colwellia psychrerythraea, Moritella marina, Photobacterium profundum, and in some marine protists thraustochytrid such as Thraustochytrium and Schizochytrium, have PUFA-PKS clustered genes homologs of the Shewanella that has raised the possibility of lateral gene transfer (105–107). These homologous PKS gene clusters from PUFA-producing microorganisms varied in gene number, gene cluster order and orientation, and enzyme domain arrangement.
Metabolic engineering has been used to successfully redesign the oleaginous microbial metabolism in order to adjust the composition and properties of microbial PUFAs (108). Metabolic engineering technology interferes with parts or the entire genome, transcriptome, metabolome proteome and lipidome of microbes; therefore, understanding these pathways is essential for modifying existing strain for PUFAs rich lipid production (109). Every oleaginous microbe has unique lipid producing capabilities, and there are a variety of strategies to alter and optimize PUFA metabolic pathway (110). Researchers all over the world manipulated PUFAs synthesis, storage, and profile pathways by overexpression of genes, blocking or knockout of genes, knockdown, suppression or activation of regulators (111, 112). In natural oleaginous microbes, metabolic engineering has made it possible to increase the production of PUFAs, and in non-oleaginous microbes, it has made it possible to reconstruct PUFA biosynthesis pathways (Table 3).
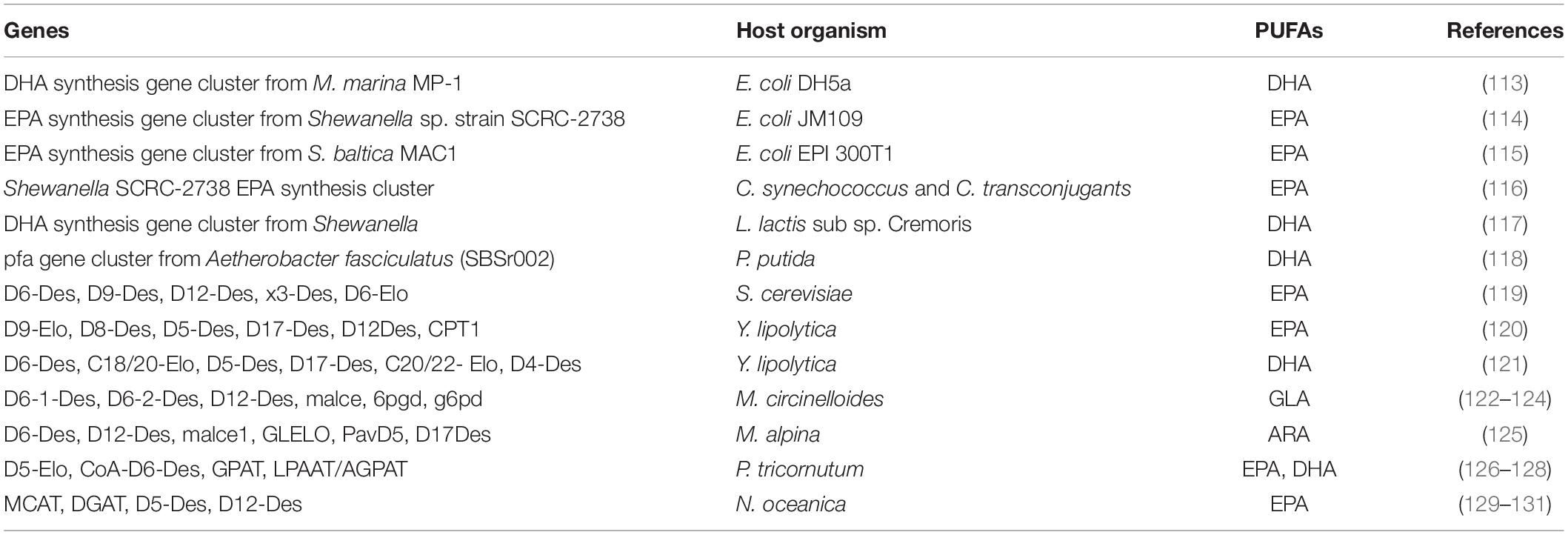
Table 3. Summary of genetic engineering of different enzymes in microbes to boost the production of PUFAs.
The discoveries of new species and early attempts to increase PUFA production have made recent advances, though they are still in the infancy stage. Studies have shown that PUFAs can be produced in heterologous bacteria such as Escherichia coli, Pseudomonas putida and lactic acid bacteria, however, the yield was very low (132). Certain barophilic and psychrophilic organisms found in the ocean depths, such as members of the Shewanella genus, have been demonstrated to be substantial PUFA producers (133). Shewanella electrodiphila MAR441 was found to produce more EPA after being supplemented with cerulenin (FAS inhibitor) (134). In Shewanella species PKS-like gene clusters were identified for EPA biosynthesis that was successfully expressed in E. coli to produce EPA (135). Heterologous expression of EPA synthesis gene cluster from Shewanella oneidensis MR-1 when expressed in E. coli XL1-Blue, low levels of EPA were reported (136). Colwellia strains, which are cold-adapted heterotrophs, produce DHA, which accounts for up to 17% of TFAs (137). DHA levels in Colwellia psychrerythraea 34H were increased with the addition of cerulenin (59). In other study DHA biosynthetic gene cluster (pfaA-D) and the pfaE gene from M. marina strain MP-1, improved recombinant DHA production in E. coli DH5a. To achieve the polyketide biosynthesis, Shewanella SCRC-2738 EPA synthesis cluster was also expressed in cyanobacteria like Cyanobacterium synechococcus and Cyanobacterial transconjugants for recombinant EPA production (116).
Several groups have reported that the biosynthetic pathways of PUFA in yeasts have been altered. Y. lipolytica, recognized for its potential to accumulate high lipid content rich in unsaturated fatty acids (138). Due to its non-pathogenic nature, Y. lipolytica has been listed in GRAS (139). Engineered oleaginous yeast, Y. lipolytica, was used to produce EPA for commercial purposes (120). The major composition of lipid content of wild-type strain of Y. lipolytica is usually PA, OA, LA, and palmitoleic acid. It was DuPont (USA) that engineered Y. lipolytica strain by overexpressing 20 desaturase genes, 8 elongase, 2 cholinephosphotransferase genes (total 30 copies codon-optimized genes) in combined with disruption of 2 genes involved in lipid metabolism and peroxisomal biogenesis factor that resulted in 56.6% EPA production from its TFA content (140). DuPont (USA) also engineered Y. lipolytica strain by overexpressing C20/22 elongase gene and Δ4 desaturase gene that resulted in the production of 18.3% DPA and 5.6% DHA (141). Overexpressing Δ6 and Δ12 desaturases from M. alpina resulted in the production of 20.2% GLA of TFAs in Y. lipolytica (121). Furthermore, the expression of bifunctional Δ12/Δ15-desaturase in Y. lipolytica resulted ALA production (142). A recent study found that the pfa cluster, which contains the pfa1, pfa2, pfa3, and ppt domains (myxobacterial gene) modified Y. lipolytica to produce high DHA (143). Recently, there has been a surge in interest in creating unusual lipids with significant industrial value. This development has been accompanied by the creation of novel synthetic biology techniques and gene editing technology for Y. lipolytica. The metabolic engineering of Y. lipolytica has produced different unusual fatty acids, such as ricinoleic acid (144, 145). The variety of lipids produced by oleaginous Y. lipolytica is expected to rise in the coming years to cope with the increasing demand.
Mortierella alpina, an oleaginous fungus, endowed with ARA found mostly in soil as a saprophyte. It should come as no surprise that strains of M. alpina are still being investigated extensively, with the goal of increasing ARA titres or broadening the PUFA range (146). M. alpina 1S-4, high accumulating ARA strain, was genetically modified to overexpress the malce1 and glelo genes, that resulted transformants to produce more ARA than the wild strain (147). A specific D17-desaturase was inserted into genome of M. alpina that resulted in an increased conversion of ARA into EPA (148). Heterologous expression of PPD17 another desaturase with high D17 selectivity (from Phytophthora parasitica) was over expressed in M. alpina that boosted the EPA 31.5% of total fatty acids (149).
Mucor circinelloides, a typical oleaginous fungus, has been extensively studied in terms of GLA production. In M. circinelloides the optimization was done to convert the upstream path-way intermediate oleic acid into GLA. Native desaturases capable of catalyzing the necessary conversion were overexpressed for this purpose. Homologous overexpression studies of two delta-6 (delta 6-2 and delta 6-1) and delta-12 desaturases in M. circinelloides increased its GLA production (122). In terms of the PUFA pathway’s downstream steps, a recent study developed the DGLA-producing mutant by heterologous expression of GLA elongase (GLELO) (150). The delta-15 desaturase gene from M. alpina was cloned and overexpressed in M. circinelloides that resulted in production of SDA (151).
Saccharomyces cerevisiae produces saturated FAs and MUFAs but no PUFAs (152). S. cerevisiae is an excellent host for eukaryotic enzymes involved in PUFA biosynthesis, such as elongases and desaturases. Recent efforts have demonstrated eicosadienoic acid, linoleic acid, and alpha-linolenic acid synthesis in recombinant S. cerevisiae, however, the production results is significantly less than oleaginous microorganisms (153). When high PUFAs are expressed in S. cerevisiae it becomes inebriated, displaying damage in their proteins, lipid peroxides, and even cell death (caspase-mediated) (154). It appears this yeast must be heavily engineered to produce significant levels of PUFAs. Low levels of EPA were also reported by co-expressing C18-PUFA specific C. elegans D6-elongase from and S. cerevisiae D6-, D5-desaturaes (155).
Microalgae have larger content of lipids in their dry cell mass with a variety of carbon length fatty acids, making them an excellent host for long chain PUFA production. Several microalgal species have been determined to be safe (GRAS), makes products of microalgae including PUFAs safe and widely acceptable for human food (156). In order increase PUFAs production, a number of lipid metabolism reaction fluxes, including upstream fatty acid generation and triacylglycerol biosynthesis must all be optimized as well. As various biochemical routes of lipid metabolism are interconnected, their regulation must be carefully managed (Figure 3).
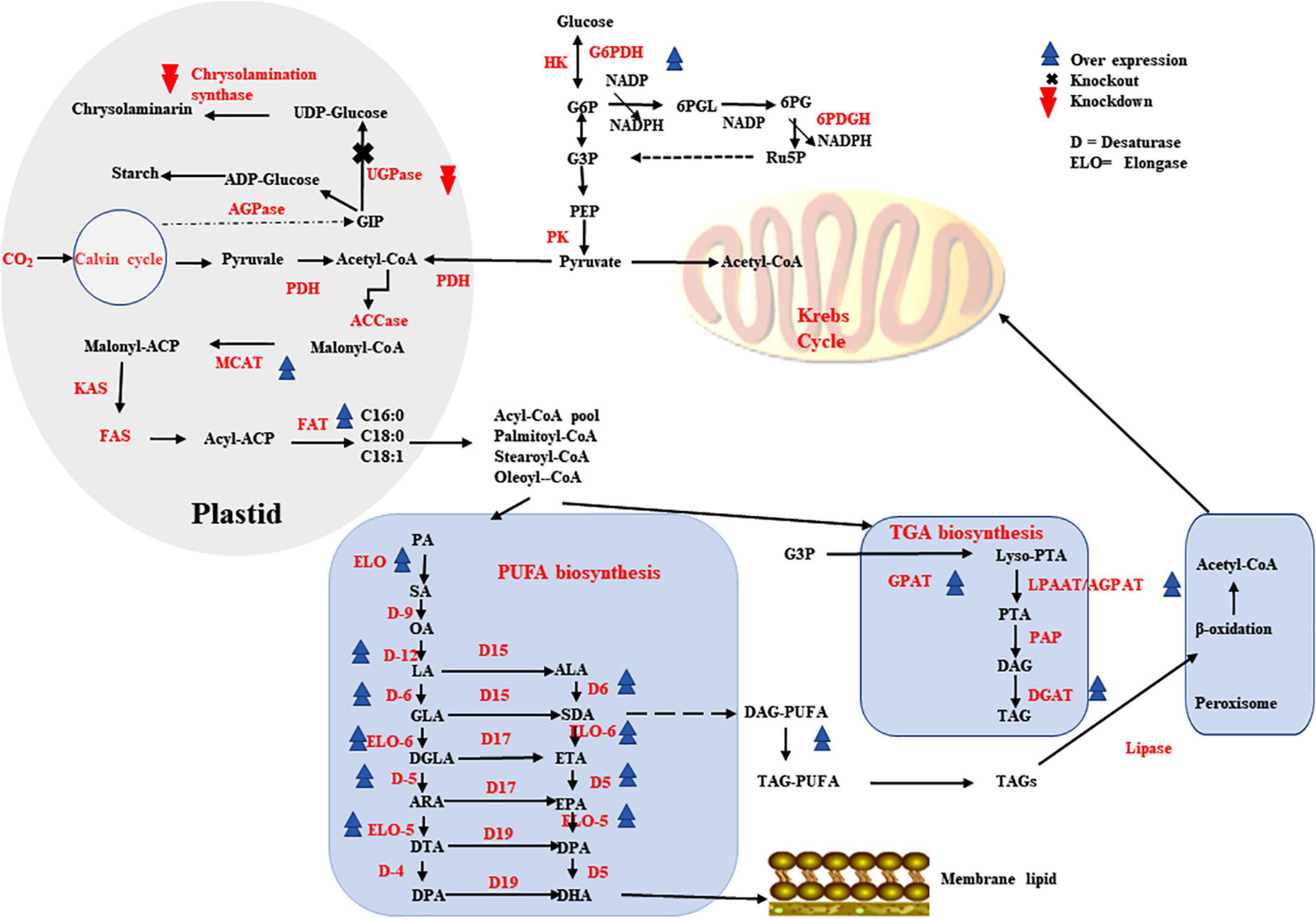
Figure 3. Metabolic engineering strategies that contributes higher production of PUFAs in higher microbes.
Malonyl CoA-acyl carrier protein transacylase enzyme aids in the formation of malonyl-ACP during the initial phase of fatty acid biosynthesis. MCAT overexpression has been shown to be very effective in higher lipid production in a variety of microalgal species for example in Nannochloropsis oceanica, overexpression resulted in increased total lipid content by 31% and EPA increased by 8% (129). Another enzyme is GPAT that catalyzes glyceraldehyde 3-phosphate conversion to lysophosphatidic acid is crucial step in TAG biosynthetic pathway. When the GPAT encoding gene was overexpressed in Phaeodactylum tricornutum, the EPA content was increased by 40% (127). The second acyltransferase enzyme in the TAG biosynthetic route is LPAAT/AGPAT, which has narrow substrate selectivity, specifically 16–18 carbon. When the gene encoding LPAAT/AGPAT was overexpressed in P. tricornutum that resulted in increased content of DHA and EPA by 1.50- and 1.55-fold, respectively (128). TAG biosynthesis downstream enzymes DGAT(s) are involved in the bondage of FAs and glycerol into TAGs and also serve as excellent target for overexpression. When the DGAT gene was overexpressed in N. oceanica, the TAG-associated PUFAs increased by 184 %, corresponding to a 4.8-fold increase in EPA (130).
It has been discovered that overexpression of FADs and/or ELOs in microalgal cells, together with other genes showed high impact on the PUFA production. In N. oceanica, overexpression of individual endogenous genes encoding Δ5- or Δ12-FAD increased EPA content by about 25% (131). Overexpression of the elo1 (encodes 6-ELO) and elo2 (encodes 5-ELO) genes in Thalassiosira pseudonana resulted in a 12% rise in EPA and a 150% increase in DHA, respectively (157). PUFAs can be improved by overexpressing 5-FAD (PtD5b) and MCAT together, as shown in P. tricornutum, which accumulated large amounts of DHA, EPA and ARA (158). Overexpression of both the OtElo5 and the glucose transporter genes in P. tricornutum allowed the transformant to grow in glucose-supplemented medium and accumulate more DHA and EPA than the wild type under phototrophic conditions, reaching 23.6 and 36.5% of total neutral lipids, respectively (92).
Phaeodactylum tricornutum lipid content increased by 24% as a result of RNA interference (RNAi)-mediated UGPase gene downregulation (159). In T. pseudonana, chrysolaminarin synthase gene knockdown increased lipid levels up to 2.4-fold (160). The favorable benefits of downregulating a putative TAG lipase-encoding gene on increased total lipid synthesis in P. tricornutum were demonstrated using RNAi technology, the EPA content in TAGs increased by 70% with slightly slower growth (161). RNAi knockdown of PEPCK enzyme (phosphoenolpyruvate carboxykinase), significantly boosted lipid accumulation by roughly 40% relative to the wild type without impairing cell biomass (162). In heterotrophic marine microalgae Aurantiochytrium sp., G6PDH-encoding gene overexpression increased the content of LC-PUFAs, especially DPA and DHA by 25 and 12.5%, respectively (163). Endogenous Malic enzyme (ME) overexpression in P. tricornutum raised NADPH content by around 80% and up to twofold the production of DHA (164).
It has been proven that metabolic engineering procedures for microorganisms are helpful in enhancing PUFAs production. During nitrogen-limiting cultivation, the fundamental difference between oleaginous and non-oleaginous microbes becomes apparent. Non-oleaginous yeasts direct carbon flow into polysaccharide synthesis, but oleaginous yeasts preferentially channel it towards lipid production, resulting in an accumulation of TAGs within distinct intracellular lipid bodies. There are still numerous gaps to fill, thus more rigorous fundamental research is required to acquire a deeper understanding of oleaginous microbial biology. Specifically, it would be of interest to discover how microbes share their metabolism of lipid, including the biosynthesis of important fatty acids, and how their growth stages and environmental factors like nutrient availabilities and cultivation mode affect the lipid metabolism. Bioprocess engineering and metabolic engineering solutions should be studied simultaneously for successful industrial scale production of microbial PUFAs.
Bioconversion of Agro-Industrial Wastes Into High-Value SCO
It has been recently brought to the attention of the scientific community that most oleaginous fungi and microalgae have the potential to grow on a variety of substrates. Agricultural raw materials used in the food industries or in industrial processing generate enormous amounts of waste each year. The availability of multiple minerals, nitrogen and carbon sources in these wastes make them appropriate as microorganism growth substrate. With the purpose of minimizing the cost of microbial lipid production, these wastes have been recommended as a substrate for oleaginous heterotrophic microbes. Furthermore, this approach should be also seen as an environmentally beneficial method of disposal of wastes (51). Oleaginous microbes that are involved in the bioconversion of agro-industrial wastes into high-value polyunsaturated fatty acids are presented in Table 4. Agricultural wastes contain a significant proportion of lignocellulosic substrates, which are mostly made of lignin, cellulose and hemicellulose, and are thus useful as carbon sources for biological transformations (165). Aquaculture wastewater is regarded as cost-effective growing media for microalgae may increase biomass yields while simultaneously removes certain nutrients from the waters, such as Nitrogen and Phosphorous (166, 167). The majority of oleaginous microbes are unable to utilize lignocellulose. The genetic manipulation of oil producing fungal strains may also allow for the direct lignocellulose conversion into SCO, a method that is previously being used in the generation of biofuels such as ethanol by using engineered yeast and fungi that are capable of assimilation of lignocellulosic materials (168). There is also another possibility to do genetic manipulation of lignocellulolytic microbes that enable them direct convert lignocellulosic substrates to lipids rich in PUFAs. Trichosporon oleaginosus, an oleaginous basidiomycete yeast well-known for its capacity to exploit xylose-containing substrates, has been modified to manufacture exceptionally LCFA, such as eicosatrienoic acid and eicosadienoic acid, which are intermediates in the production of the EPA and DHA (169).
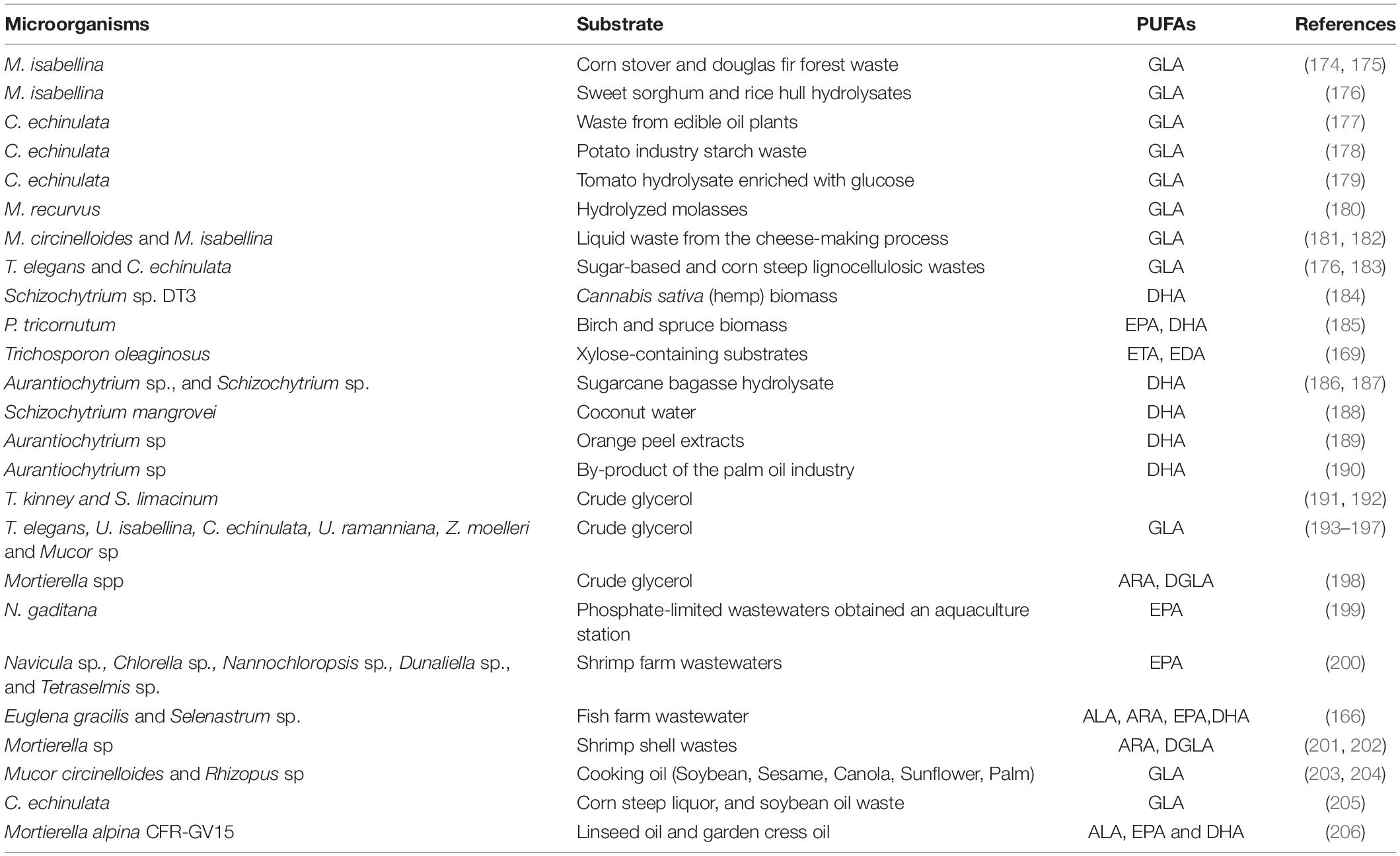
Table 4. Summary of bioconversion of agro-industrial wastes into high-value single cell oil by oleaginous microbes.
Finding suitable substrates is one of the most important factors in large-scale SCO production, and oleaginous microorganisms can ferment both hydrophilic and hydrophobic substrates. However, lipid fermentation on hydrophilic and hydrophobic substrates follows two distinct bio-pathways: “de novo” and “ex novo.” It is worth noting that both pathway of lipid fermentation have advantages: “de novo” lipid fermentation typically produces a large amount of lipid, whereas “ex novo” lipid fermentation can transform the extracellular and intracellular lipid composition to meet the needs of the food or chemical industries. The ability of microorganisms to use glucose, xylose, arabinose, etc. as carbon sources is highly desired in order to increase the efficiency of biomass and lipid production, while an important drawback associated with the lipid fermentative conversion of sugar-based residues is the production of other metabolites such as intracellular polysaccharides, extracellular citric acid. For example under nitrogen-limited conditions, M. isabellina ATHUM 2935 cultivated on glucose/xylose blends, demonstrated remarkably different physiological and biochemical patterns related to the sugar used as substrate. When glucose-rich media were used as carbon sources, significant lipid production occurred, while endopolysaccharides and polyols (mannitol and xylitol) were also produced in varying amounts depending on the substrate composition (170).
There have been reports of oleaginous microorganisms assimilation of hydrophobic substrates such as esters, TAGs, FAs, sterols, etc. via distinct pathway namely ex novo lipid synthesis (170). Hydrophobic materials acquired from an industrial waste stream, waste cooking oils, effluents from dairy and butter producing enterprises, and waste fish oils can all be used as hydrophobic feedstock for biomass and lipid production by oleaginous microbes (171). Y. lipolytica has been widely investigated in an effort to understand how hydrophobic substrates are utilized by the organism (172). The active transport of hydrophobic substrates into the cell is facilitated by secreted lipases, therefore only the microorganisms capable of producing lipases can incorporate free FAs. Once inside the cell, free FAs can be used for energy or they can be integrated into lipidic structures for storage. Fat biomodification by oleaginous microbes can be used to tailor the FAs profiles of hydrophobic substrates into value-added oils (173). It is common for hydrophobic substrates to form layers on the surface of the water, which prevents the oleaginous yeast from accessing them during fermentation experiments. Despite this, some oleaginous yeast can produce extracellular emulsifiers that can reduce the size of hydrophobic substrates and improve their miscibility in water. To make hydrophobic substrates more accessible to oleaginous microorganisms, researchers sometimes added emulsifiers like Tween 80, Span 80, Tween 60, and OP-10 to the aqueous medium. The addition of emulsifiers to the fermenting medium, on the other hand, raises the overall production cost and has an impact on the final lipid production because it is frequently dissolved in organic solvents, interfering with the estimation of residual hydrophobic substrates.
Approaches That Leads Novel Sources of Microbial Polyunsaturated Fatty Acids
Tapping New Microbial Sources
Based on DNA analysis and mathematical modeling, it is widely acknowledged that just a small fraction of microbial diversity has been studied, and that even the majority of microbial species have yet to be isolated and tested for chemical diversity (72, 207). Unusual habitats surveys have produced numerous novel microbes with important fatty acids in the past, hence unknown microbial species occupying rare habitats with various environmental restrictions are regarded resources for PUFA production (72, 208–211). Underexplored ecosystems include hyperarid, high-saline environments desert, forest as well as unique microbes that coexist with higher species in symbiotic/mutualistic interactions. With this strategy, a new PUFA microbial source has already been discovered (75, 212). The oceans, which cover more than 70% of the Earth’s surface, are a rich source of microorganisms with different morphological, physiological, and chemical diversity. Considering that just 7–8% of oceans are made up of coastal areas, the deep-sea biosphere, which extends from the seashore to depths of >10,000 m, contains 95% of the ocean’s microorganism biomass and is a potential source for novel microbial metabolites (213). The sampling of deep ocean sediments remains one of the obstacles in harnessing new source of microorganisms. Access to the deep ocean is generally dependent on conventional oceanographic processes that are not specifically developed for microbiological research, which excludes huge areas of the ocean floor from sampling range (214). The Antarctic marine environment is home to cold-adapted prokaryotic and eukaryotic species with unique characters that allow them to survive in saltwater at a constant low temperature. A number of Antarctic microorganisms have shown an increase in fatty acid unsaturation with decreasing growing temperature (76). It is necessary for deep-sea bacteria to thrive in a psychrophilic and piezophilic habitat for EPA to take effect (210). Shewanella livingstonensis Ac10, for example, is a psychrotrophic bacteria isolated from Antarctic saltwater that produces a considerable amount of EPA (Kawamoto et al. 2009). There is a marine bacterium known as Shewanella piezotolerans WP3 that has been identified as psychrotolerant and piezotolerant (75). It was found in sediment from the western Pacific Ocean at a depth of 1914 m, where it thrived at temperatures and pressures as high as 20 MPa. EPA is found in the membrane of S. piezotolerans cells (215, 216). At a depth of 5110 meters, the Ryukyu Trench was home to the discovery of Shewanella violacea strain DSS12. At 8 °C, and 30 MPa, it thrives under a moderately piezophilic environment. Even at 0.1 MPa, it can grow at 8°C, S. violacea DSS12 cells have a 15% EPA membrane content (216, 217). Several marine bacterial cultures capable of producing ω-3 FAs have been isolated in recent years, and the metabolic pathways involved in their synthesis have been identified. It has been recognized that the majority of known microbial producers are gamma-Proteobacteria, specifically those belonging to the genera Shewanella, Photobacteria, Colwellia, Vibrio, Psychromonas, and Moritella (76, 218). Some isolates associated with Psychroserpens and Flexibacter in the Flavobacteria class have also showed the ability to synthesize ω-3 LC-PUFAs (75). Several thraustochytrid strains have been isolated from seawater (219–221). Thraustochytrids are well-known for producing important ω-3 PUFAs like EPA, and DHA (222). Thraustochytrids may store lipids in excess of 50% of their dry weight, with DHA accounting for more than 25% of TFAs (212).
Cultivating Rare Microbes
The importance of rare microorganisms in microbial communities is still a topic of debate. Scientists estimate that there are at least 10 million different kinds of microorganisms in the world, and DNA data shows that just 1% of those microorganisms can be easily cultivated in a lab (223, 224). This is due to the fact that many rare microbes are slow-growing, some may have specialized requirements. Many rare microbes that require special media have been isolated by changing media conditions, such as utilizing seawater-based isolation medium to allow marine organisms to grow (225). Signaling molecules like homoserine lactones, cAMP and pyruvate been demonstrated to increase the number of microorganisms from various habitats and assist uncultured strains recover (226). This type of approach was found to be very successful in isolating microbes that produced various bioactive secondary metabolites. However, as far as microbes that produce PUFA are concerned, there has been no such report till date. But this approach can’t be underestimated in exploring the microbial world for the production of PUFAs.
High-Throughput Screening
High-throughput screening approaches use automated screening and large-scale data analysis to speed up the discovery of new oleaginous microorganism sources. HTS is very useful in the screening process of strain libraries for microbial metabolites (227, 228). Because of the miniaturization of fermentation technologies, it is now possible to test a large number of strains under carefully regulated conditions. The flexibility to work with a variety of strains and targeted models is one of the benefits of HTS, as it saves both time and resources. Till date, a number of oleaginous yeasts have been investigated, including Basidiomycota and Ascomyta, despite the fact that the majority of genera and species have yet to be tested for oil content (229–231). One significant factor to remember is that yeast taxonomy is always changing, therefore when determining whether a yeast species is oleaginous or not, special attention should be paid to identification of species. To quantify lipids generated by microorganisms, standard technologies such as HPLC, GC, TLC, and MS are available. Although these methods are highly reliable and credible, they require expensive and heavy equipment, making large-scale yeast screens time-consuming and difficult (232, 233). A standard lipid extraction method must be developed for all oleaginous microorganisms in order to avoid the use of harmful organic solvents and other drawbacks associated with previous methods for quantification in various biological models. Numerous approaches for quick screening have been proposed in the past like fluorimetric methods using Sudan black (234), Nile red (235), and Nile blue (236) and, presently the use of FTIR (237). Following this basic screening, more robust and thorough methods for confirming and validating the results should be applied. HTS tools for assessing fatty acid composition in oleaginous yeast and microalgae species are currently being used, with promising results (238).
Co-culturing and Elicitation
Under normal laboratory fermentation conditions, many putative gene clusters can go unnoticed or unexpressed making the production of microbial PUFAs via established conventional routes difficult. The major research on microbial oil has been done on single cell cultures. New methods for activating silent genes that involve the co-culture (two or more microorganisms are cultivated together in the same environment) are gaining popularity. In the co-culturing method, producer strain released the products by the activation of molecules or precursors secreted by inducer strain. Because of the interactions between diverse cultures that present in most natural environment, this system has been regarded as an effective model. Mostly microbes are frequently found in intimate connection with one another; they live and grow in the same biocoenosis for as long as source of nutrient are accessible. As a result, microbial interactions between cultures might be exploited and reproduced at the laboratory scale. This is not a new concept; for decades, mixed cultures have played a key role in wastewater treatment, creating biomass and bioactive chemicals production (239–241). Elicitation involves presenting microbial cells with a variety of signals, and the use of these so-called ‘elicitors’ has been shown to be effective in the production of various new metabolites (242, 243). When microorganisms detect these external signals, they produce discrete reactions that result in a changed metabolite profile, and if induced chemicals are precisely targeted, new structures are expected.
To attain much better lipid productivity, scientists are using a co-cultivation technique that attempts to reconcile the discrepancy between biomass productivity and lipid content. Various studies have been conducted to improve lipid productivity by encouraging the accumulation of total biomass and lipid output through the use of a co-culture strategy with various microbial species. In general, research on this method has been described for algae species e.g., the co-culture of C. sorokiniana and Chlorella vulgaris with Azospirillum brasilense increased not only lipid, but also variety of fatty acids, (244). Many other species have been reported to have high growth coupled with other microbial species, such Rhodotorula glutinis and Ambrosiozyma cicatricose (245–247) and Monoraphidium sp FXY10 (248, 249), Dostalek found that the co-culture of R. toruloides and Saccharomycopsis fibuligera in starch media resulted in a greater lipid and biomass content (250).
Genome-Mining Approaches
The rapid move from single-gene to entire-genome analysis was made possible by advances in sequencing technology, which provide yet another significant platform for revealing microorganisms’ potential (242). Diverse putative gene clusters remain silent under various conditions, making it challenging to discover their entire metabolic potential. HTS can aid in the discovery of silent gene clusters in microbial genomes, as well as the subsequent characterization of cryptic products using heterologous gene expression or gene knockdown experiments combined (251, 252). When a species’ genetic information is insufficient, such as in many “non-model” organisms, de novo genome assembly makes it possible to do functional genomics research on them. It was discovered that the PKS pathway, which is responsible for the manufacture of DHA, requires sufficient NADPH and acetyl-CoA, and that 30 genes were anticipated to be possible targets for DHA overproduction in Schizochytrium limacinum SR21 (253). Two newly identified Thraustochytrids strains, SW8 and Mn4, had their genomes dramatically devoured by secreted carbohydrate-active enzymes, as revealed by comparative genomic research. The FAS and PKS routes of PUFA generation were found to be incomplete (254). Thraustochytrium sp. 26185’s genome sequence and analysis also revealed the presence of anaerobic and aerobic PUFAs metabolic pathways. But, the aerobic route lacked a crucial gene for stearate delta-9 desaturase. Although the genome of NP10 was found to be extremely similar to that of numerous strains in the S. griseus clade, comparative research targeted at identifying potential FA synthases assisted in the discovery of a prospective gene cluster. Following gene inactivation and complementation, it was discovered that this cluster was involved in the manufacture of a variety of FFAs in the NP10 organism (255). This genome mining strategy resulted in the putative pfa gene cluster identification in a number of S. cellulosum strains. A fatty acid methylesters analysis by GC-MS revealed that in these strains’ PUFA synthesis is limited to eicosadienoic acid (EDA) and linoleic acid (LA) (256). Additionally, from the genus Aetherobacter, myxobacterial isolates have been found to be prolific makers of LC- PUFAs. Using FAME, these strains were found to produce AA, EPA, and DHA (257, 258). Anaerobic polyunsaturated fatty acid (PUFA) production via PKS-like and FAS-like synthases was discovered in many terrestrial myxobacteria of the Sorangiineae suborder (259). The LC- PUFAs EPA and ARA were also found in the Enhygromyxa salina SHK-1T member of Nannocystineae (260, 261). Genome mining of the closely related strains E. salina SWB005, E. salina SWB007 E. and salina DSM 15201, using publically available genome sequences also indicated pfa gene clusters presence that was identical to known myxobacterial pfa clusters (262).
Genomic Analysis
A thorough picture of lipid metabolism in oleaginous microorganisms was provided by genome analysis, route mapping, and major lipid profiling. It will be critical to understand how gene expression is regulated during lipogenesis in various oleaginous microorganisms, as well as how it is coordinated between the creation of precursor molecules like pyruvate and acetyl-CoA, the production of the reductant NADPH, and the synthesis of lipids.
The M. alpina genome is 38.38 Mb in size and contains a significant number of gene duplications. More than half of its 12,796 gene models and 60% of the estimated lipogenesis pathway’s genes are part of multigene families. M. alpina’s fatty acid synthase is a single polypeptide that contains all of the catalytic domains required for fatty acid whereas this enzyme is composed of two polypeptides in many fungi. According to phylogenetic study based on genes involved in lipid metabolism, oleaginous fungus may have gained their lipogenic capacity throughout evolution following the divergence of Ascomycota, Basidiomycota, Chytridiomycota, and Mucoromycota (229). FADS12 and FADS15 from M. alpina catalyze the production of omega-6 and omega-3 polyunsaturated fatty acids, respectively. More than half of the total fat in M. alpina was composed of AA, which is synthesized at a much higher rate than EPA. Because there is just one gene for each desaturase in the M. alpina genome, this difference in capacity cannot be explained by gene duplication. (229). However, it is probable that the difference is due to the expression levels or catalytic activity of FADS12 and FADS15, as forced expression of FADS15 in M. alpina 1S-4 significantly boosts EPA synthesis (263).
Comparative genomics approach facilitates the identification of multiple differently expressed genes between distinct strains of the same microorganism (264). The genome of M. circinelloides CBS 277.49, a low lipid generating strain, was compared to M. circinelloides WJ11, a high lipid generating strain. The similarities between CBS 277.49 and WJ11 suggested that they are similar in terms of gene identity and gene order. The WJ11 G+C content and size of genome assembly were found to be about 39% and 35.4 Mb, respectively. it has also been revealed that the genomes of the two strains have numerous homologous regions and these regions were mostly co-linear. The number of genes for lipid accumulation enzymes was compared via reorganization of lipid metabolism and central carbon routes (265). There are two genes encoding Δ9 desaturase in both strains. As M. circinelloides is a GLA-producing filamentous fungus, as expected, in both strains, there were two genes encoding fatty acid delta-12 desaturase (Δ12) to convert oleic acid to linoleic acid and one gene encoding fatty acid delta-6 desaturase (Δ6) to convert linoleic acid into γ-linolenic acid (265). The majority of enzymes in the PPP pathway were encoded by the same number of genes in WJ11 and CBS 277.49, however, in WJ11, G6PDH was encoded by four genes, but in CBS it was encoded by three genes. More genes encoding G6PDH may indicate that WJ11 has more opportunity to create NADPH for fatty acid biosynthesis (265). Some enzymes in the TCA cycle, such as succinyl-CoA ligase (SUCLG), succinate dehydrogenase (SDH), and fumarate hydratase (FH), were encoded by fewer genes in WJ11 as compared to CBS strain, indicating a more active TCA cycle in CBS that in turn has the inverse relationship to fatty acid accumulation (265).
Safety, Toxicity and Cost of Microbial Polyunsaturated Fatty Acids
Triacylglycerides and sterols, which are identical to those found in plant and animal oils, are the major components of microbial oils, with the exception of bacterial oils. Microbial oils, however, are considered new foods due to their origin, and certain regulatory hurdles must be passed before they can be utilized as food. Microbial oils have been examined for their safety, nutritional value, and clinical usefulness for human use. It is critical to study the producing organism while analyzing the safety of any microbial oil. There should not have a history of creating hazardous metabolites, nor should it be capable of infecting animals or harming the environment. In the absence of any doubt, pathogenic organisms should not be taken into account, as there is always the possibility of live organisms escaping from a production facility to infect surroundings. Oils may thus be shown to have extremely minimal non-lipid content and so cannot contain any substantial amount of any element that is likely to be dangerous (266). When oil was introduced from a new source, such as the first SCO from M. circinelloides, M. alpina, C. cohnii, and Schizochytrium sp which all represented “new” sources of oil at the time of their introduction, regulatory authorities required complete dossiers of evidence for the safety of these oils. In fact, the oil was purer than other plant oils, as herbicide and pesticide residues were virtually missing from the microbial oil. An important factor in this decision was that the producing organism had already received GRAS (Generally Recognized As Safe) certification. Microbial oils have been shown to be safe, but this evidence must be evaluated by various regulatory bodies around the world: The FDA in the United States; the EU ‘Regulation on novel foods and novel food ingredients’; the Canadian Food and Drugs Regulation; the Australian/New Zealand Food Standards Code; etc. These regulatory bodies are ultimately responsible for approving the sale of new foods and nutraceuticals. In the case of M. circinelloides, preliminary toxicity tests were carried out with brine shrimps, followed by 90-day animal feeding studies with several rats and other animals. All studies were fully successful, and the data was subsequently given to the UK Advisory Committee on Novel Foods and Processes for review and approval. There were no problems to the oil being sold in the UK (266). Since it has now been two decades since microbial oils have been consumed by infants, adults, and a wide range of animals, including fish and poultry. There has been no substantiated report that there has been a problem with their consumption, and the SCOs’ manufacturers and distributors have no reason to doubt their products’ complete safety and reliability. The safety of DHASCO™ (high DHA microbial oil) and ARASCO™ (high ARA microbial oil) in newborn formulae has been shown in numerous non-clinical and clinical investigations.
The cost of microbial oil is mostly determined by the species used for cultivation, cell biomass and the concentration of lipids within cells (267–271). Table 5 compares the amount of accumulated oil in some selected oleaginous microbes and plants. Oil production of microbes is generally expressed in kg/m3/year, whereas plant oil productivity is expressed in kg/ha/year. Jones et al. found that growing C. curvatus in a 30–50 m3 fermenter gives the same amount of oil as 150 palm trees in a 1-ha area in a year (271).
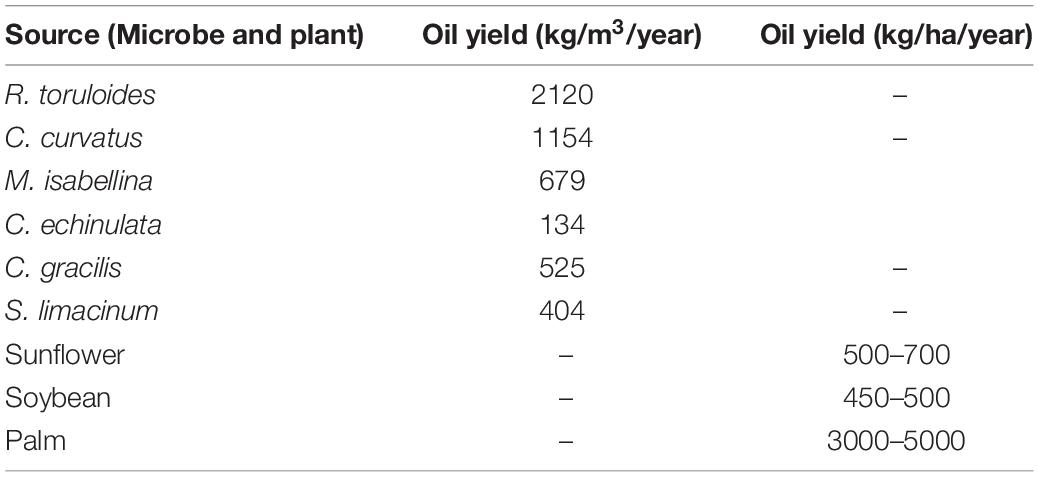
Table 5. Lipid productivity comparison of some selected oleaginous microbes and plants. Microbial oil is generally expressed in kg/m3/year, whereas plant oil productivity is expressed in kg/ha/year.
Only a few techno-economic studies have addressed the prices of microbial oil that ranges between $1.72 and $5.9/kg as compared to plant oils which are sold between $ 0.5 and 1.9/kg (270). 60 to 75 % of the total price of the microbial oil is related to the cost of the substrates needed by microbes to produce the lipids. Using an effective strain and low-cost substrate can therefore help to increase the profitability of single-cell oil production.
Future Perspectives and Concluding Remarks
There is a limited supply of polyunsaturated fatty acids on the commercial market, so scientists around the world are attempting to discover and develop new strains of organisms with essential PUFA profiles. Microbes have gained a lot of attention for their potential when it comes to producing PUFAs, but only a small percentage of them have been cultured so far. The combination of genomic platforms with high-throughput screening and fermentation studies could reveal hidden microbial’ PUFAs producing potential. It’s worth noting that oil production via fermentative procedures using microorganisms is quite costly. Greater attention must be paid to the extraction of oils from novel or genetically engineered species. The high cost of microbial oil production must be solved by utilizing low-cost carbon and other nutrients for growth, as well as a low-cost harvesting approach for biomass recovery and a superior lipids extraction procedure. Bio refinery techniques for PUFAs production, for total exploitation of microbial oils, will, in addition to other pathways, address the challenges related with PUFAs production cost. We should keep looking for novel PUFA-rich microorganisms from nature that can make a contribution to this sector. Multidisciplinary initiatives integrating microbiologists, molecular biologists, and natural product chemists are necessary to expedite PUFA generation from microorganisms.
Author Contributions
AS and YZ performed literature research and wrote manuscript draft. WY and HM performed fact checking and finalized table and figures. YS designed and revised the final draft. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (Grants Nos. 31972851 and 31670064), TaiShan Industrial Experts Program (No. tscy20160101), and Natural Science Foundation of Jiangsu Province (Grant No. BK20190265).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
PUFAs, polyunsaturated fatty acids; SCO, single-cell oil; HTS, high-throughput screening; FAs, fatty acids; C, carbon; MUFAs, monounsaturated fatty acids; ω, omega; LC, liquid chromatography; GC, gas chromatography; EI, electrospray ionization; MS, mass spectrometry; NMR, nuclear magnetic resonance; MALDI, matrix-assisted laser desorption/ionization; ALA, α -linolenic acid; ARA, arachidonic acid; DGLA, dihomo- γ -linolenic acid; DHA, docosahexaenoic acid; DPA, docosapentaenoic acid; EPA, eicosapentaenoic acid; GLA, γ -linolenic acid; MA, mead acid; SDA, stearidonic acid; TAGs, triacylglycerides; PKS, polyketide synthase; GRAS, generally regarded as safe; PA, palmitic acid; OA, oleic acid; LA, linoleic acid; TFAs, total fatty acids; MCAT, malonyl CoA-acyl carrier protein transacylase; GPAT, Glyceraldehyde 3-phosphate acyltransferase; LPAAT, Lysophosphatidate acyltransferase; DGAT(s), Diacylglycerol acyltransferases; PEPCK, phosphoenolpyruvate carboxykinase; G6PDH, glucose-6-phosphate dehydrogenase; LCFA, long chain fatty acids; HPLC, high-performance liquid chromatography; TLC, thin-layer chromatography; FTIR, fourier transform Infrared spectroscopy.
Footnotes
References
1. Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Murphy RC, et al. Comprehensive classification system for lipids1. J Lipid Res. (2005) 46:839–61. doi: 10.1194/jlr.E400004-JLR200
2. De Carvalho CC, Caramujo MJ. The various roles of fatty acids. Molecules. (2018) 23:2583. doi: 10.3390/molecules23102583
3. Sikorski ZZ, Kolakowska A. Chemical and Functional Properties of Food Lipids. Boca Raton, FL: CRC press (2010). doi: 10.1201/9781003040392
4. Efsa Panel on Dietetic Products N, Journal Aje. Scientific opinion on dietary reference values for fats, including saturated fatty acids, polyunsaturated fatty acids, monounsaturated fatty acids, trans fatty acids, and cholesterol. (2010) 8:1461. doi: 10.2903/j.efsa.2010.1461
5. Jovanovic S, Dietrich D, Becker J, Kohlstedt M, Wittmann C. Microbial production of polyunsaturated fatty acids—high-value ingredients for aquafeed, superfoods, and pharmaceuticals. Curr Opin Biotechnol. (2021) 69:199–211. doi: 10.1016/j.copbio.2021.01.009
6. Meyer BJ, Mann NJ, Lewis JL, Milligan GC, Sinclair AJ, Howe PR. Dietary intakes and food sources of omega-6 and omega-3 polyunsaturated fatty acids. Lipids. (2003) 38:391–8. doi: 10.1007/s11745-003-1074-0
7. Bajpai PK, Bajpai P, Ward OP. Optimisation of culture conditions for production of eicosapentaenoic acid by Mortierella elongate NRRL 5513. J Ind Microbiol Biotechnol. (1992) 9:11–7. doi: 10.1007/BF01576363
8. Köfeler HC, Fauland A, Rechberger GN, Trötzmüller M. Mass spectrometry based lipidomics: an overview of technological platforms. Metabolites. (2012) 2:19–38. doi: 10.3390/metabo2010019
9. Zárate R, El Jaber-Vazdekis N, Tejera N, Pérez JA, Rodríguez C. Significance of long chain polyunsaturated fatty acids in human health. Clin Transl Med. (2017) 6:25. doi: 10.1186/s40169-017-0153-6
10. Cerone M, Smith TK. A Brief journey into the history of and future sources and uses of fatty acids. Front Nutr. (2021) 8:570401. doi: 10.3389/fnut.2021.570401
11. Mhlongo S, Ezeokoli OT, Roopnarain A, Ndaba B, Sekoai PT, Habimana O, et al. The potential of single-cell oils derived from filamentous fungi as alternative feedstock sources for biodiesel production. Front microbiol. (2021) 12:637381. doi: 10.3389/fmicb.2021.63738
12. Hu C, van der Heijden R, Wang M, van der Greef J, Hankemeier T, Xu G. Analytical strategies in lipidomics and applications in disease biomarker discovery. J Chromatogr B. (2009) 877:2836–46. doi: 10.1016/j.jchromb.2009.01.038
13. Ochsenreither K, Glück C, Stressler T, Fischer L, Syldatk CJ. Production strategies and applications of microbial single cell oils. Fron Microbiol. (2016) 7:1539. doi: 10.3389/fmicb.2016.01539
14. Lopes da Silva T, Moniz P, Silva C, Reis A. The dark side of microalgae biotechnology: a heterotrophic biorefinery platform directed to ω-3 rich lipid production. Microorganisms. (2019) 7:670. doi: 10.3390/microorganisms7120670
15. Elrazak A, Ahmed AA. Production of Polyunsaturated Fatty Acids from Marine Microorganisms. Doctoral dissertation. Newcastle upon Tyne: Newcastle University (2012).
16. Colombo SM, Rodgers TF, Diamond ML, Bazinet RP, Arts MT. Projected declines in global DHA availability for human consumption as a result of global warming. Ambio. (2020) 49:865–80. doi: 10.1007/s13280-019-01234-6
17. Shahidi F, Ambigaipalan P. Omega-3 polyunsaturated fatty acids and their health benefits. Annu Rev Food Sci Technol. (2018) 9:345–81. doi: 10.1146/annurev-food-111317-095850
18. Rey C, Delpech J, Madore C, Nadjar A, Greenhalgh AD, Amadieu C, et al. Dietary n-3 long chain PUFA supplementation promotes a pro-resolving oxylipin profile in the brain. Brain Behav. (2019) 76:17–27. doi: 10.1016/j.bbi.2018.07.025
19. Cholewski M, Tomczykowa M, Tomczyk M. A comprehensive review of chemistry, sources and bioavailability of omega-3 fatty acids. Nutrients. (2018) 10:1662. doi: 10.3390/nu10111662
20. Bentsen H. Dietary polyunsaturated fatty acids, brain function and mental health. Microb Ecol Health Dis. (2017) 28(Sup. 1):1281916. doi: 10.1080/16512235.2017.1281916
21. Yanai H, Masui Y, Katsuyama H, Adachi H, Kawaguchi A, Hakoshima M, et al. An improvement of cardiovascular risk factors by omega-3 polyunsaturated fatty acids. J Clin Med Res. (2018) 10:281–9. doi: 10.14740/jocmr3362w
22. Djuricic I, Calder PC. Beneficial outcomes of omega-6 and omega-3 polyunsaturated fatty acids on human health: an update for 2021. Nutrients. (2021) 13:2421. doi: 10.3390/nu13072421
23. Nordøy A, Marchioli R, Arnesen H, Videbæk J. n- 3 Polyunsaturated fatty acids and cardiovascular diseases: to whom, how much, preparations. Lipids. (2001) 36:S127–9. doi: 10.1007/s11745-001-0695-7
24. Metherel A, Armstrong J, Patterson A, Stark KD. Assessment of blood measures of n-3 polyunsaturated fatty acids with acute fish oil supplementation and washout in men and women. Prostaglandins Leukot Essent Fatty Acids. (2009) 81:23–9. doi: 10.1016/j.plefa.2009.05.018
25. Lagarde M, Burtin M, Sprecher H, Dechavanne M, Renaud S. Potentiating effect of 5, 8, 11-eicosatrienoic acid on human platelet aggregation. Lipids. (1983) 18:291–4. doi: 10.1007/BF02534704
26. Phang M, Garg ML, Sinclair AJ. Inhibition of platelet aggregation by omega-3 polyunsaturated fatty acids is gender specific—Redefining platelet response to fish oils. Prostaglandins Leukot. Essent Fatty Acids. (2009) 81:35–40. doi: 10.1016/j.plefa.2009.05.001
27. Das UN. Long-chain polyunsaturated fatty acids in the growth and development of the brain and memory. Nutrition. (2003) 19:62. doi: 10.1016/s0899-9007(02)00852-3
28. Das UN. From bench to the clinic: γ-linolenic acid therapy of human gliomas. Prostaglandins Leukot Essent Fatty Acids. (2004) 70:539–52. doi: 10.1016/j.plefa.2003.12.001
29. Sayegh F, Elazzazy A, Bellou S, Moustogianni A, Elkady AI, Baeshen MN, et al. Production of polyunsaturated single cell oils possessing antimicrobial and anticancer properties. Ann Microbiol. (2016) 66:937–48. doi: 10.1007/s13213-015-1176-0
30. Alakhras R, Bellou S, Fotaki G, Stephanou G, Demopoulos NA, Papanikolaou S, et al. Fatty acid lithium salts from Cunninghamella echinulata have cytotoxic and genotoxic effects on HL-60 human leukemia cells. Eng Life Sci. (2015) 15:243–53. doi: 10.1002/elsc.201400208
31. Saelao S, Kanjana-Opas A, Kaewsuwan S. Optimization of biomass and arachidonic acid production by Aureispira maritima using response surface methodology. JAOCS. (2011) 88:619–29. doi: 10.1007/s11746-010-1710-y
32. Jain T. Fatty acid composition of oilseed crops: a Review. Innov Food Sci Emerg Technol. (2020) 13:147–53. doi: 10.1007/978-981-15-2556-8_13
33. Desbois AP, Lawlor KC. Antibacterial activity of long-chain polyunsaturated fatty acids against Propionibacterium acnes and Staphylococcus aureus. Mar Drugs. (2013) 11:4544–57. doi: 10.3390/md11114544
34. Das UN. Arachidonic acid and other unsaturated fatty acids and some of their metabolites function as endogenous antimicrobial molecules: a review. J Adv Res. (2018) 11:57–66. doi: 10.1016/j.jare.2018.01.001
35. Meyer B, Tsivis E, Howe PC, Tapsell L, Calvert G. Polyunsaturated fatty acid content of foods: differentiating between long and short chain omega-3 fatty acids. Food Australia. (1999) 51:81–95. doi: 10.1111/fwb.13492
36. Salem N Jr., Eggersdorfer MJ. Is the world supply of omega-3 fatty acids adequate for optimal human nutrition? Curr Opin Clin Nutr Metab Care. (2015) 18:147–54. doi: 10.1097/MCO.0000000000000145
37. Ratledge C. Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie. (2004) 86:807–15. doi: 10.1016/j.biochi.2004.09.017
38. del Rosario González-Baró Ma Pollero RJ. Fatty acid metabolism of Macrobrachium borellii: dietary origin of arachidonic and eicosapentaenoic acids. Comp Biochem Physiol. (1998) 119:747–52. doi: 10.1016/S1095-6433(98)01028-9
39. Papanikolaou S, Seraphim P, Papanikolaou S. Oleaginous yeasts: biochemical events related with lipid synthesis and potential biotechnological applications. J Ferment. (2012) 1:1–3. doi: 10.4172/2167-7972.1000e103
40. Dewick PM. Medicinal Natural Products: A Biosynthetic Approach. Hoboken, NJ: John Wiley & Sons (2002).
41. Ratledgea C. Micro-organismes producteurs de lipides. OCL. (2013) 20:D602. doi: 10.1051/ocl/2013029
42. Čertík M, Balteszov L, Šajbidor J. Lipid formation and γ-linolenic acid production by Mucorales fungi grown on sunflower oil. Braz J Microbiol. (1997) 25:101–5. doi: 10.1046/j.1472-765X.1997.00173.x
43. Gema H, Kavadia A, Dimou D, Tsagou V, Komaitis M, Aggelis G. Production of γ-linolenic acid by Cunninghamella echinulata cultivated on glucose and orange peel. Appl Microbiol Biotechnol. (2002) 58:303–7. doi: 10.1007/s00253-001-0910-7
45. Amano N, Shinmen Y, Akimoto K, Kawashima H, Amachi T. Chemotaxonomic significance of fatty acid composition in the genus Mortierella (Zygomycetes, Mortierellaceae). Mycotaxon. (1992) 44:257–65.
46. Shimizu S, Jareonkitmongkol S. Mortierella Species (Fungi): Production of C 20 Polyunsaturated Fatty Acids. In: Medicinal and Aromatic Plants VIII. Berlin: Springer (1995). p. 308–25. doi: 10.1007/978-3-662-08612-4_17
47. Radwan SS. Sources of C 20-polyunsaturated fatty acids for biotechnological use. Appl Microbiol Biotechnol. (1991) 35:421–30. doi: 10.1007/BF00169743
48. Eroshin V, Dedyukhina E, Chistyakova T, Zhelifonova V, Botast RJ. Studies on arachidonic acid production by Mortierella fungi: a microbiological method for selecting arachidonic acid producers. Microbiology (New York, NY). (1996) 65:26–31.
49. Nakajima T. Microbial production and purification of ω 6 polyunsaturated fatty acids. In: A Sinclair, R Gibson editors. Essential Fatty Acids and Eicosanoids. Champaign, IL: American Oil Chemists’ Society (1993). p. 57–64.
50. Garcia RO, Reichenbach H, Ring MW, Müller R, microbiology e. Phaselicystis flava gen. nov., sp. nov., an arachidonic acid-containing soil myxobacterium, and the description of Phaselicystidaceae fam. nov. Int J Syst Evol Microbiol. (2009) 59:1524–30. doi: 10.1099/ijs.0.003814-0
51. Kothri M, Mavrommati M, Elazzazy AM, Baeshen MN, Moussa TA, Aggelis G. Microbial sources of polyunsaturated fatty acids (PUFAs) and the prospect of organic residues and wastes as growth media for PUFA-producing microorganisms. FEMS Microbiol Lett. (2020) 367:fnaa028. doi: 10.1093/femsle/fnaa028
52. Makri A, Bellou S, Birkou M, Papatrehas K, Dolapsakis NP, Bokas D, et al. Lipid synthesized by micro-algae grown in laboratory-and industrial-scale bioreactors. Eng Life Sci. (2011) 11:52–8. doi: 10.1002/elsc.201000086
53. Suh S-S, Kim SJ, Hwang J, Park M, Lee T-K, Kil E-J, et al. Fatty acid methyl ester profiles and nutritive values of 20 marine microalgae in Korea. Asian Pac J Trop Med. (2015) 8:191–6. doi: 10.1016/S1995-7645(14)60313-8
55. Leman J. Oleaginous microorganisms: an assessment of the potential. Adv Appl Microbiol. (1997) 43:195–243. doi: 10.1016/s0065-2164(08)70226-0
56. Nettleton JA. Omega-3 Fatty Acids and Health. Boston, MA: Springer (1995). p. 64–76. doi: 10.1007/978-1-4615-2071-9_2
57. Vazhappilly R, Chen F. Eicosapentaenoic acid and docosahexaenoic acid production potential of microalgae and their heterotrophic growth. JAOCS. (1998) 75:393–7. doi: 10.1007/s11746-998-0057-0
58. Kendrick A, Ratledge C. Lipids of selected molds grown for production of n- 3 and n- 6 polyunsaturated fatty acids. Lipids. (1992) 27:15–20. doi: 10.1007/BF02537052
59. Wan X, Peng Y-F, Zhou X-R, Gong Y-M, Huang F-H, Moncalián G. Effect of cerulenin on fatty acid composition and gene expression pattern of DHA-producing strain Colwellia psychrerythraea strain 34H. Microb Cell Factories. (2016) 15:1–13. doi: 10.1186/s12934-016-0431-9
60. Nakahara T, Yokochi T, Higashihara T, Tanaka S, Yaguchi T, Honda D. Production of docosahexaenoic and docosapentaenoic acids bySchizochytrium sp. isolated from Yap Islands. JAOCS. (1996) 73:1421–6.
61. Shimiziu S, Kawashima H, Shinmen Y, Akimoto K, Yamada H. Production of eicosapentaenoic acid byMortierella fungi. JAOCS. (1988) 65:1455–9. doi: 10.1007/BF02898307
62. Weete JD. Enhancement of C_ polyunsaturated fatty acid production in Pythium ultimun. In: DJ Kyle, C Ratledge editors. Industrial Applications of Single Cell Oils. Champaign, IL: American Oil Chemists’ Society (1992). p. 98–117. doi: 10.1201/9781439821855.ch6
63. Boswell KD. SCO production by fermentative microalgae. In: DJ Kyle, C Ratledge editors. Industrial Applications of Single Cell Oils. Champaign, IL: American Oil Chemists’ Society (1992). p. 274–86. doi: 10.1201/9781439821855.ch15
64. Yazawa K. Pruduction of eicosapentaenoic acid from marine bacteria. In: DJ Kyle, C Ratledge editors. Industrial Applications of Single Cell Oils. Champaign, IL: American Oil Chemists’ Society (1992). p. 29–51. doi: 10.1201/9781439821855.ch3
65. Oliw E, Sprecher H, Hamberg M. Biosynthesis of a novel prostaglandin. Δ17-PGE1, in the ram. Acta Physiol Scand. (1986) 127:45–9. doi: 10.1111/j.1748-1716.1986.tb07874.x
66. Kawashima H, Kamada N, Sakuradani E, Jareonkitmongkol S, Akimoto K, Shimizu S. Production of 8, 11, 14, 17-cis-eicosatetraenoic acid by Δ5 desaturase-defective mutants of an arachidonic acid-producing fungus, Mortierella alpina. JAOCS. (1997) 74:455–9. doi: 10.1007/s11746-997-0106-0
68. Yokochi T. Lipid productivity and fatty acid composition of fungal class Zygomycetes containing gamma-linolenic acid. J Natl Chem Lab Ind. (1992) 87:463–70.
69. Čertík M, Berhan SS, Šajbidor J. Lipid production and fatty acid composition of selected strains belonging to Mucorales. Acta Biotechnologica. (1993) 13:193–6. doi: 10.1002/abio.370130219
70. Certik M, Sajbidor J. Variability of fatty acid composition in Mucor and Rhizopus strains and its dependence on submersed and surface growth. Microbios. (1996) 85:151–60.
71. Cohen Z. Production of polyunsaturated fatty acids (EPA, ARA, and GLA) by the microalgae Porphyridium and Spirulina. In: DJ Kyle, C Ratledge editors. Industrial Applications of Single Cell Oils. Champaign, IL: American Oil Chemists’ Society (1992). p. 243–73. doi: 10.1201/9781439821855.ch14
72. Shah AM, Mohamed H, Zhang Z, Song Y. Isolation, characterization and fatty acid analysis of Gilbertella persicaria DSR1: a potential new source of high value single-cell oil. Biomass Bioenergy. (2021) 151:106156. doi: 10.1016/j.biombioe.2021.106156
73. Kawashima H, Nishihara M, Hirano Y, Kamada N, Akimoto K, Konishi K, et al. Production of 5, 8, 11-eicosatrienoic acid (Mead acid) by a (Delta) 6 desaturation activity-enhanced mutant derived from a (Delta) 12 desaturase-defective mutant of an arachidonic acid-producing fungus, Mortierella alpina 1S-4. Appl. Environ Microbiol. (1997) 63:1820–5. doi: 10.1128/aem.63.5.1820-1825.1997
74. DeLong EF, Yayanos AA., microbiology e. Biochemical function and ecological significance of novel bacterial lipids in deep-sea procaryotes. Appl Environ Microbiol. (1986) 51:730–7. doi: 10.1128/aem.51.4.730-737.1986
75. Moi IM, Leow ATC, Ali MSM, Rahman RNZRA, Salleh AB, Sabri S. Polyunsaturated fatty acids in marine bacteria and strategies to enhance their production. Appl Microbiol Biotechnol. (2018) 102:5811–26. doi: 10.1007/s00253-018-9063-9
76. Freese E, Rütters H, Köster J, Rullkötter J, Sass H. Gammaproteobacteria as a possible source of eicosapentaenoic acid in anoxic intertidal sediments. Microb Ecol. (2009) 57:444–54. doi: 10.1007/s00248-008-9443-2
77. Allen EE, Facciotti D, Bartlett DH. Monounsaturated but not polyunsaturated fatty acids are required for growth of the deep-sea bacterium Photobacterium profundum SS9 at high pressure and low temperature. Appl Environ Microbiol. (1999) 65:1710–20. doi: 10.1128/AEM.65.4.1710-1720.1999
78. Fang J, Kato C, Sato T, Chan O, McKay D. Biosynthesis and dietary uptake of polyunsaturated fatty acids by piezophilic bacteria. Comp Biochem Physiol. (2004) 137:455–61. doi: 10.1016/j.cbpc.2004.01.009
79. Nogi Y, Hosoya S, Kato C, Horikoshi K. Psychromonas hadalis sp. nov., a novel piezophilic bacterium isolated from the bottom of the Japan Trench. Int J Sys Evol Microbiol. (2007) 57:1360–4. doi: 10.1099/ijs.0.64933-0
80. Bharathiraja B, Sridharan S, Sowmya V, Yuvaraj D, Praveenkumar R. Microbial oil–a plausible alternate resource for food and fuel application. Bioresour Technol. (2017) 233:423–32. doi: 10.1016/j.biortech.2017.03.006
81. Laoteng K, Čertík M, Cheevadhanark S. Mechanisms controlling lipid accumulation and polyunsaturated fatty acid synthesis in oleaginous fungi. Chem Zvesti. (2011) 65:97–103. doi: 10.2478/s11696-010-0097-4
82. Ward OP, Singh A. Omega-3/6 fatty acids: alternative sources of production. Process Biochem. (2005) 40:3627–52. doi: 10.1016/j.procbio.2005.02.020
83. Ratledge C. Microbial oils: an introductory overview of current status and future prospects. OCL. (2013) 20:D602. doi: 10.1051/ocl/2013029
84. Miller KK, Alper HS. Yarrowia lipolytica: more than an oleaginous workhorse. Appl Microbiol Biotechnol. (2019) 103:9251–62. doi: 10.1007/s00253-019-10200-x
85. Oliver L, Dietrich T, Marañón I, Villarán MC, Barrio RJ. Producing omega-3 polyunsaturated fatty acids: a review of sustainable sources and future trends for the EPA and DHA market. Resources. (2020) 9:148. doi: 10.3390/resources9120148
86. Tanaka T, Yabuuchi T, Maeda Y, Nojima D, Matsumoto M, Yoshino T. Production of eicosapentaenoic acid by high cell density cultivation of the marine oleaginous diatom Fistulifera solaris. Bioresour Technol. (2017) 245:567–72. doi: 10.1016/j.biortech.2017.09.005
87. Wen ZY, Chen F. Application of statistically-based experimental designs for the optimization of eicosapentaenoic acid production by the diatom Nitzschia laevis. In: F Chen, Y Jiang editors. Algae and their Biotechnological Potential. (Vol. 75), Dordrecht: Springer (2001). p. 159–69. doi: 10.1002/bit.1175
88. Martins DA, Custódio L, Barreira L, Pereira H, Ben-Hamadou R, Varela J, et al. Alternative sources of n-3 long-chain polyunsaturated fatty acids in marine microalgae. Mar Drugs. (2013) 11:2259–81. doi: 10.3390/md11072259
89. Chen C-Y, Lee M-H, Dong C-D, Leong YK, Chang J-S. Enhanced production of microalgal lipids using a heterotrophic marine microalga Thraustochytrium sp. BM2 Biochem Eng J. (2020) 154:107429. doi: 10.1016/j.bej.2019.107429
90. Shi H, Luo X, Wu R, Yue X. Production of eicosapentaenoic acid by application of a delta-6 desaturase with the highest ALA catalytic activity in algae. Microb Cell Factories. (2018) 17(1):1–15. doi: 10.1186/s12934-018-0857-3
91. Chen C-Y, Nagarajan D, Cheah WY. Eicosapentaenoic acid production from Nannochloropsis oceanica CY2 using deep sea water in outdoor plastic-bag type photobioreactors. Bioresour Technol. (2018) 253:1–7. doi: 10.1016/j.biortech.2017.12.102
92. Hamilton ML, Powers S, Napier JA, Sayanova O. Heterotrophic production of omega-3 long-chain polyunsaturated fatty acids by trophically converted marine diatom Phaeodactylum tricornutum. Mar Drugs. (2016) 14:53. doi: 10.3390/md14030053
93. Diao J, Song X, Guo T, Wang F, Chen L, Zhang W. Cellular engineering strategies toward sustainable omega-3 long chain polyunsaturated fatty acids production: State of the art and perspectives. Biotechnol Adv. (2020) 40:107497. doi: 10.1016/j.biotechadv.2019.107497
94. Norashikin MN, Loh SH, Aziz A, San Cha T. Metabolic engineering of fatty acid biosynthesis in Chlorella vulgaris using an endogenous omega-3 fatty acid desaturase gene with its promoter. Algal Res. (2018) 31:262–75. doi: 10.1016/j.algal.2018.02.020
95. Metz JG, Roessler P, Facciotti D, Levering C, Dittrich F, Lassner M, et al. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science. (2001) 293:290–3. doi: 10.1126/science.1059593
96. Hashimoto K, Yoshizawa AC, Okuda S, Kuma K, Goto S, Kanehisa M. The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomess?? J. Lipid Res. (2008) 49:183–91. doi: 10.1194/jlr.M700377-JLR200
97. Pereira SL, Leonard AE, Mukerji P. Recent advances in the study of fatty acid desaturases from animals and lower eukaryotes. Prostaglandins Leukot Essent Fatty Acids. (2003) 68:97–106. doi: 10.1016/s0952-3278(02)00259-4
98. García-Maroto F, Garrido-Cárdénas JA, Rodríguez-Ruiz J, Vilches-Ferrón M, Adam AC, Polaina J, et al. Cloning and molecular characterization of the Δ6-desaturase from two Echium plant species: production of GLA by heterologous expression in yeast and tobacco. Lipids. (2002) 37:417–26. doi: 10.1007/s1145-002-0910-6
99. Sayanova O, Smith MA, Lapinskas P, Stobart AK, Dobson G, Christie WW, et al. Expression of a borage desaturase cDNA containing an N-terminal cytochrome b5 domain results in the accumulation of high levels of Δ6-desaturated fatty acids in transgenic tobacco. Proc Natl Acad Sci U.S.A. (1997) 94:4211–6. doi: 10.1073/pnas.94.8.4211
100. Piispanen R, Saranpää P. Neutral lipids and phospholipids in Scots pine (Pinus sylvestris) sapwood and heartwood. Tree Physiology. (2002) 22:661–6. doi: 10.1093/treephys/22.9.661
101. Leonard AE, Pereira SL, Sprecher H, Huang YS. Elongation of long-chain fatty acids. Prog Lipid Res. (2004) 43:36–54. doi: 10.1016/S0163-7827(03)00040-7
102. Dittrich F, Zajonc D, Hühne K, Hoja U, Ekici A, Greiner E, et al. Fatty acid elongation in yeast: biochemical characteristics of the enzyme system and isolation of elongation-defective mutants. Eur J Biochem. (1998) 252:477–85. doi: 10.1046/j.1432-1327.1998.2520477.x
103. Tehlivets O, Scheuringer K, Kohlwein SD. Fatty acid synthesis and elongation in yeast. Biochim Biophys Acta Biomembr. (2007) 1771:255–70. doi: 10.1016/j.bbalip.2006.07.004
104. Yazawa K. Production of eicosapentaenoic acid from marine bacteria. Lipids. (1996) 31:S297–300. doi: 10.1007/BF02637095
105. Allen EE, Bartlett DH. Structure and regulation of the omega-3 polyunsaturated fatty acid synthase genes from the deep-sea bacterium Photobacterium profundum strain SS9. Microbiology. (2002) 148:1903–13. doi: 10.1099/00221287-148-6-1903
106. Tanaka M, Ueno A, Kawasaki K, Yumoto I, Ohgiya S, Hoshino T, et al. Isolation of clustered genes that are notably homologous to the eicosapentaenoic acid biosynthesis gene cluster from the docosahexaenoic acid-producing bacterium Vibrio marinus strain MP-1. Biotechnol Lett. (1999) 21:939–45. doi: 10.1023/A:1005601606929
107. Methé BA, Nelson KE, Deming JW, Momen B, Melamud E, Zhang X, et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc Natl Acad Sci U.S.A. (2005) 102:10913–8. doi: 10.1073/pnas.0504766102
108. Yuan S-F, Alper HS. Metabolic engineering of microbial cell factories for production of nutraceuticals. Microb Cell Factories. (2019) 18:1–11. doi: 10.1186/s12934-019-1096-y
109. Pomraning KR, Kim Y-M, Nicora CD, Chu RK, Bredeweg EL, Purvine SO, et al. Multi-omics analysis reveals regulators of the response to nitrogen limitation in Yarrowia lipolytica. BMC Genom. (2016) 17:138. doi: 10.1186/s12864-016-2471-2
110. Mohamed H, Naz T, Yang J, Shah AM, Nazir Y, Song Y. Recent molecular tools for the genetic manipulation of highly industrially important Mucoromycota fungi. J Fungi. (2021) 7:1061. doi: 10.3390/jof7121061
111. Ostergaard S, Olsson L, Nielsen J. Metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol Rev. (2000) 64:34–50. doi: 10.1128/MMBR.64.1.34-50.2000
112. Ji X-J, Huang H. Engineering microbes to produce polyunsaturated fatty acids. Trends Biotechnol. (2019) 37:344–6. doi: 10.1016/j.tibtech.2018.10.002
113. Orikasa Y, Nishida T, Yamada A, Yu R, Watanabe K, Hase A, et al. Recombinant production of docosahexaenoic acid in a polyketide biosynthesis mode in Escherichia coli. Biotechnol. (2006) 28:1841–7. doi: 10.1007/s10529-006-9168-6
114. Orikasa Y, Yamada A, Yu R, Ito Y, Nishida T, Yumoto I, et al. Characterization of the eicosapentaenoic acid biosynthesis gene cluster from Shewanella sp. strain SCRC-2738. Cell Mol Biol (Noisy-le-grand) (2004) 50:625–30.
115. Amiri-Jami M, Griffiths MW. Recombinant production of omega-3 fatty acids in Escherichia coli using a gene cluster isolated from Shewanella baltica MAC1. J Appl Microbiol. (2010) 109:1897–905. doi: 10.1111/j.1365-2672.2010.04817.x
116. Takeyama H, Takeda D, Yazawa K, Yamada A, Matsunaga T. Expression of the eicosapentaenoic acid synthesis gene cluster from Shewanella sp. in a transgenic marine cyanobacterium, Synechococcus sp. Microbiology (Reading, Engl.). (1997) 143:2725–31. doi: 10.1099/00221287-143-8-2725
117. Amiri-Jami M, LaPointe G, Griffiths MW. Engineering of EPA/DHA omega-3 fatty acid production by Lactococcus lactis subsp. cremoris MG1363. Appl Microbiol Biotechnol. (2014) 98(7):3071–80. doi: 10.1007/s00253-013-5381-0
118. Gemperlein K, Zipf G, Bernauer HS, Müller R, Wenzel SC. Metabolic engineering of Pseudomonas putida for production of docosahexaenoic acid based on a myxobacterial PUFA synthase. Metab. (2016) 33:98–108. doi: 10.1016/j.ymben.2015.11.001
119. Tavares S, Grotkjær T, Obsen T, Haslam RP, Napier JA, Gunnarsson N. Metabolic engineering of Saccharomyces cerevisiae for production of eicosapentaenoic acid, using a novel Δ5-desaturase from Paramecium tetraurelia. Appl Environ Microbiol. (2011) 77:1854–61. doi: 10.1128/AEM.01935-10
120. Xue Z, Sharpe PL, Hong S-P, Yadav NS, Xie D, Short DR. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat Biotechnol. (2013) 31:734–40. doi: 10.1038/nbt.2622
121. Damude HG, Zhang H, Farrall L, Ripp KG, Tomb J-F, Hollerbach D, et al. Identification of bifunctional Δ12/ω3 fatty acid desaturases for improving the ratio of ω3 to ω6 fatty acids in microbes and plants. Proc Natl Acad Sci U S A. (2006) 103:9446–51. doi: 10.1073/pnas.0511079103
122. Zhang Y, Luan X, Zhang H, Garre V, Song Y, Ratledge C. Improved γ-linolenic acid production in Mucor circinelloides by homologous overexpressing of delta-12 and delta-6 desaturases. Microb Cell Factories. (2017) 16:1–9. doi: 10.1186/s12934-017-0723-8
123. Zhang Y, Adams IP, Ratledge C. Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology (Reading, Engl.) (2007) 153:2013–25. doi: 10.1099/mic.0.2006/002683-0
124. Tang X, Chen H, Gu Z, Zhang H, Chen YQ, Song Y, et al. Role of g 6 pdh and leuB on Lipid accumulation in Mucor circinelloides. J Agric Food Chem. (2020) 68:4245–51. doi: 10.1021/acs.jafc.9b08155
125. Kikukawa H, Sakuradani E, Ando A, Shimizu S, Ogawa J. Arachidonic acid production by the oleaginous fungus Mortierella alpina 1S-4: A review. J Adv Res. (2018) 11:15–22. doi: 10.1016/j.jare.2018.02.003
126. Hamilton ML, Haslam RP, Napier JA, Sayanova O. Metabolic engineering of Phaeodactylum tricornutum for the enhanced accumulation of omega-3 long chain polyunsaturated fatty acids. Metab. (2014) 22:3–9. doi: 10.1016/j.ymben.2013.12.003
127. Niu YF, Wang X, Hu D-X, Balamurugan S, Li D-W, Yang WD, et al. Molecular characterization of a glycerol-3-phosphate acyltransferase reveals key features essential for triacylglycerol production in Phaeodactylum tricornutum. Biotechnol Biofuels. (2016) 9:1–11. doi: 10.1186/s13068-016-0478-1
128. Balamurugan S, Wang X, Wang H-L, An C-J, Li H, Li DW, et al. Occurrence of plastidial triacylglycerol synthesis and the potential regulatory role of AGPAT in the model diatom Phaeodactylum tricornutum. Biotechnol Biofuels. (2017) 10:1–14. doi: 10.1186/s13068-017-0786-0
129. Chen JW, Liu WJ, Hu DX, Wang X, Balamurugan S, Alimujiang A, et al. Identification of a malonyl CoA-acyl carrier protein transacylase and its regulatory role in fatty acid biosynthesis in oleaginous microalga Nannochloropsis oceanica. Appl Biochem Biotechnol. (2017) 64:620–6. doi: 10.1002/bab.1531
130. Xin Y, Lu Y, Lee YY, Wei L, Jia J, Wang Q, et al. Producing designer oils in industrial microalgae by rational modulation of co-evolving type-2 diacylglycerol acyltransferases. Mol Plant. (2017) 10:1523–39. doi: 10.1016/j.molp.2017.10.011
131. Poliner E, Pulman JA, Zienkiewicz K, Childs K, Benning C, Farré EM. A toolkit for Nannochloropsis oceanica CCMP 1779 enables gene stacking and genetic engineering of the eicosapentaenoic acid pathway for enhanced long-chain polyunsaturated fatty acid production. Plant Biotechnol. (2018) 16:298–309. doi: 10.1111/pbi.12772
132. Kannan N, Rao AS, Nair A. Microbial production of omega-3 fatty acids: an overview. J Appl Microbiol. (2021) 131:2114–30. doi: 10.1111/jam.15034
133. Dailey FE, McGraw JE, Jensen BJ, Bishop SS, Lokken JP, Dorff KJ, et al. The microbiota of freshwater fish and freshwater niches contain omega-3 fatty acid-producing Shewanella species. Appl Environ Microbiol. (2016) 82:218–31. doi: 10.1128/AEM.02266-15
134. Zhang J, Burgess JG. Enhanced eicosapentaenoic acid production by a new deep-sea marine bacterium Shewanella electrodiphila MAR441T. PLoS One. (2017) 12:e0188081. doi: 10.1371/journal.pone.0188081
135. Orikasa Y, Ito Y, Nishida T, Watanabe K, Morita N, Ohwada T, et al. Enhanced heterologous production of eicosapentaenoic acid in Escherichia coli cells that co-express eicosapentaenoic acid biosynthesis pfa genes and foreign DNA fragments including a high-performance catalase gene, vktA. Biotechnol Lett. (2007) 29:803–9. doi: 10.1007/s10529-007-9310-0
136. Lee S-J, Jeong Y-S, Kim D-U, Seo J-W, Hur BK. Eicosapentaenoic acid (EPA) biosynthetic gene cluster of Shewanella oneidensis MR-1: cloning, heterologous expression, and effects of temperature and glucose on the production of EPA in Escherichia coli. Biotechnol Bioprocess Eng. (2006) 11:510–5. doi: 10.1007/BF02932075
137. Techtmann SM, Fitzgerald KS, Stelling SC, Joyner DC, Uttukar SM, Harris AP, et al. Colwellia psychrerythraea strains from distant deep sea basins show adaptation to local conditions. Front Environ Sci. (2016) 4:33. doi: 10.3389/fenvs.2016.00033
138. Zeng SY, Liu HH, Shi TQ, Song P, Ren LJ, Huang H, et al. Recent advances in metabolic engineering of Yarrowia lipolytica for lipid overproduction. Eur J Lipid Sci Technol. (2018) 120:1700352. doi: 10.1002/ejlt.201700352
139. Groenewald M, Boekhout T, Neuvéglise C, Gaillardin C, van Dijck PW, Wyss MJ. Yarrowia lipolytica: safety assessment of an oleaginous yeast with a great industrial potential. Crit Rev Microbiol. (2014) 40:187–206. doi: 10.3109/1040841X.2013.770386
140. Chuang LT, Chen D-C, Nicaud JM, Madzak C, Chen YH, Huang Y-S. Co-expression of heterologous desaturase genes in Yarrowia lipolytica. N Biotechnol. (2010) 27:277–82. doi: 10.1016/j.nbt.2010.02.006
141. Damude HG, Macool DJ, Picataggio SK, Ragghianti JJ, Seip JE, Xue Z, et al. Docosahexaenoic acid producing strains of Yarrowia lipolytica. United States patent US. (2009) 7:286.
142. Cordova LT, Alper HS. Production of α-linolenic acid in Yarrowia lipolytica using low-temperature fermentation. Appl Microbiol Biotechnol. (2018) 102:8809–16. doi: 10.1007/s00253-018-9349-y
143. Gemperlein K, Dietrich D, Kohlstedt M, Zipf G, Bernauer HS, Wittmann C, et al. Polyunsaturated fatty acid production by Yarrowia lipolytica employing designed myxobacterial PUFA synthases. Nat Commun. (2019) 10:1–12. doi: 10.1038/s41467-019-12025-8
144. Jia YL, Wang LR, Zhang ZX, Gu Y, Sun XM. Recent advances in biotechnological production of polyunsaturated fatty acids by Yarrowia lipolytica. Crit Rev Food Sci Nutr. (2021):1–15. doi: 10.1080/10408398.2021.1937041
145. Szczepańska P, Hapeta P, Lazar Z. Advances in production of high-value lipids by oleaginous yeasts. Crit Rev Biotechnol. (2021) 42:1–22. doi: 10.1080/07388551.2021.1922353
146. Rayaroth AC, Tomar RS, Mishra RK. Arachidonic acid synthesis in Mortierella alpina: Origin, evolution and advancements. Proc Natl Acad Sci U.S.A. (2017) 87:1053–66. doi: 10.1007/s40011-016-0714-2
147. Sakuradani E, Ando A, Shimizu S, Ogawa J. Metabolic engineering for the production of polyunsaturated fatty acids by oleaginous fungus Mortierella alpina 1S-4. J. Biosci Bioeng. (2013) 116:417–22. doi: 10.1016/j.jbiosc.2013.04.008
148. Ge C, Chen H, Mei T, Tang X, Chang L, Gu Z, et al. Application of a ω-3 desaturase with an arachidonic acid preference to eicosapentaenoic acid production in Mortierella alpina. Front Bioeng Biotechnol. (2018) 5:89. doi: 10.3389/fbioe.2017.00089
149. Tang X, Chen H, Mei T, Ge C, Gu Z, Zhang H, et al. Characterization of an omega-3 desaturase from Phytophthora parasitica and application for eicosapentaenoic acid production in Mortierella alpina. Front Microbiol. (2018) 9:1878. doi: 10.3389/fmicb.2018.01878
150. Khan MAK, Yang J, Hussain SA, Zhang H, Liang L, Garre V, et al. Construction of DGLA producing cell factory by genetic modification of Mucor circinelloides. Microb Cell Factories. (2019) 18:64. doi: 10.1186/s12934-019-1110-4
151. Khan M, Kabir A, Yang J, Hussain SA, Zhang H, Garre V, et al. Genetic modification of Mucor circinelloides to construct stearidonic acid producing cell factory. Int J Mol Sci. (2019) 20:1683. doi: 10.3390/ijms20071683
152. Yazawa H, Iwahashi H, Kamisaka Y, Kimura K, Uemura H. Production of polyunsaturated fatty acids in yeast Saccharomyces cerevisiae and its relation to alkaline pH tolerance. Yeast. (2009) 26:167–84. doi: 10.1002/yea.1659
153. Guo M, Chen G, Chen J, Zheng M. Identification of a long-chain fatty acid elongase from Nannochloropsis sp. involved in the biosynthesis of fatty acids by heterologous expression in Saccharomyces cerevisiae. J Ocean Univ China. (2019) 18:1199–206. doi: 10.1007/s11802-019-3946-y
154. Johansson M, Chen X, Milanova S, Santos C, Petranovic D. PUFA-induced cell death is mediated by Yca1p-dependent and-independent pathways, and is reduced by vitamin C in yeast. FEMS Yeast Res. (2016) 16:fow007. doi: 10.1093/femsyr/fow007
155. Beaudoin F, Michaelson LV, Hey SJ, Lewis MJ, Shewry PR, Sayanova O, et al. Heterologous reconstitution in yeast of the polyunsaturated fatty acid biosynthetic pathway. Proc Natl Acad Sci U S A. (2000) 97:6421–6. doi: 10.1073/pnas.110140197
156. Arterburn LM, Oken HA, Hall EB, Hamersley J, Kuratko CN, Hoffman JP. Algal-oil capsules and cooked salmon: nutritionally equivalent sources of docosahexaenoic acid. J Am Diet Assoc. (2008) 108:1204–9. doi: 10.1016/j.jada.2008.04.020
157. Cook O, Hildebrand M. Enhancing LC-PUFA production in Thalassiosira pseudonana by overexpressing the endogenous fatty acid elongase genes. J Appl Phycol. (2016) 28:897–905. doi: 10.1007/s10811-015-0617-2
158. Wang X, Liu YH, Wei W, Zhou X, Yuan W, Balamurugan S, et al. Enrichment of long-chain polyunsaturated fatty acids by coordinated expression of multiple metabolic nodes in the oleaginous microalga Phaeodactylum tricornutum. J Agric Food Chem. (2017) 65:7713–20. doi: 10.1021/acs.jafc.7b02397
159. Zhu B-H, Shi H-P, Yang G-P, Lv N-N, Yang M, Pan K-H. Silencing UDP-glucose pyrophosphorylase gene in Phaeodactylum tricornutum affects carbon allocation. N Biotechnol. (2016) 33:237–44. doi: 10.1016/j.nbt.2015.06.003
160. Hildebrand M, Manandhar-Shrestha K, Abbriano R. Effects of chrysolaminarin synthase knockdown in the diatom Thalassiosira pseudonana: implications of reduced carbohydrate storage relative to green algae. Algal Res. (2017) 23:66–77. doi: 10.1016/j.algal.2017.01.010
161. Li X, Pan Y, Hu H. Identification of the triacylglycerol lipase in the chloroplast envelope of the diatom Phaeodactylum tricornutum. Algal Res. (2018) 33:440–7. doi: 10.1016/j.algal.2018.06.023
162. Yang J, Pan Y, Bowler C, Zhang L, Hu H. Knockdown of phosphoenolpyruvate carboxykinase increases carbon flux to lipid synthesis in Phaeodactylum tricornutum. Algal Res. (2016) 15:50–8. doi: 10.1016/j.algal.2016.02.004
163. Cui GZ, Ma Z, Liu YJ, Feng Y, Sun Z, Cheng Y, et al. Overexpression of glucose-6-phosphate dehydrogenase enhanced the polyunsaturated fatty acid composition of Aurantiochytrium sp. SD116 Algal Res. (2016) 19:138–45. doi: 10.1016/j.algal.2016.08.005
164. Zhu B-H, Zhang R-H, Lv N-N, Yang G-P, Wang Y-S, Pan KH. The role of malic enzyme on promoting total lipid and fatty acid production in Phaeodactylum tricornutum. Front Plant Sci. (2018) 9:826. doi: 10.3389/fpls.2018.00826
165. De Bhowmick G, Sarmah AK, Sen R. Zero-waste algal biorefinery for bioenergy and biochar: a green leap towards achieving energy and environmental sustainability. Sci Total Environ. (2019) 650:2467–82. doi: 10.1016/j.scitotenv.2018.10.002
166. Tossavainen M, Lahti K, Edelmann M, Eskola R, Lampi A-M, Piironen V, et al. Integrated utilization of microalgae cultured in aquaculture wastewater: wastewater treatment and production of valuable fatty acids and tocopherols. J Appl Phycol. (2019) 31:1753–63. doi: 10.1007/s10811-018-1689-6
167. Guo Z, Liu Y, Guo H, Yan S, Mu J. Microalgae cultivation using an aquaculture wastewater as growth medium for biomass and biofuel production. Res J Environ Sci. (2013) 25:S85–8. doi: 10.1016/S1001-0742(14)60632-X
168. Elkins JG, Raman B, Keller M. Engineered microbial systems for enhanced conversion of lignocellulosic biomass. Curr Opin Biotechnol. (2010) 21:657–62. doi: 10.1016/j.copbio.2010.05.008
169. Görner C, Redai V, Bracharz F, Schrepfer P, Garbe D, Brück T. Genetic engineering and production of modified fatty acids by the non-conventional oleaginous yeast Trichosporon oleaginosus ATCC 20509. Green Chem. (2016) 18:2037–46. doi: 10.1039/C5GC01767J
170. Gardeli C, Athenaki M, Xenopoulos E, Mallouchos AK, Koutinas AA, Aggelis G, et al. Lipid production and characterization by Mortierella (Umbelopsis) isabellina cultivated on lignocellulosic sugars. J Appl Microbiol. (2017) 123:1461–77. doi: 10.1111/jam.13587
171. Carsanba E, Papanikolaou S, Erten H. Production of oils and fats by oleaginous microorganisms with an emphasis given to the potential of the nonconventional yeast Yarrowia lipolytica. Crit Rev Biotechnol. (2018) 38:1230–43. doi: 10.1080/07388551.2018.1472065
172. Beopoulos A, Chardot T, Nicaud JM. Yarrowia lipolytica: a model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie. (2009) 91:692–6. doi: 10.1016/j.biochi.2009.02.004
173. Ghazani SM, Marangoni AG. Microbial lipids for foods. Trends Food Sci Technol. (2021) 119:593–607. doi: 10.1016/j.tifs.2021.10.014
174. Fang H, Zhao C, Chen S. Single cell oil production by Mortierella isabellina from steam exploded corn stover degraded by three-stage enzymatic hydrolysis in the context of on-site enzyme production. Bioresour Technol. (2016) 216:988–95. doi: 10.1016/j.biortech.2016.06.051
175. Ivančić Šantek M, Santek B, Beluhan S. Chapter Production of Microbial Lipids from Lignocellulosic Biomass. London: InTechOpen (2018). doi: 10.5772/intechopen.74013
176. Economou CN, Aggelis G, Pavlou S, Vayenas DV. Single cell oil production from rice hulls hydrolysate. Bioresour Technol. (2011) 102:9737–42. doi: 10.1016/j.biortech.2011.08.025
177. Van der Merwe R, Badenhorst J, Britz T. Fungal treatment of an edible-oil-containing industrial effluent. World J Microbiol Biotechnol. (2005) 21:947. doi: 10.1007/s11274-004-6962-y
178. Wang H, Deng Z, Zhou S, Zhang X. Using potato starch waste water to produce GLA with Cunninghamella echinulata. China Oil Fat. (2007) 32:49–51.
179. Fakas S, Papanikolaou S, Galiotou-Panayotou M, Komaitis M, Aggelis G. Organic nitrogen of tomato waste hydrolysate enhances glucose uptake and lipid accumulation in Cunninghamella echinulata. J app. microbiol. (2008) 105:1062–70. doi: 10.1111/j.1365-2672.2008.03839.x
180. Li N, Deng Z-N, Qin Y-L, Chen C-L, Liang ZQ. Production of polyunsaturated fatty acids by Mucor recurvus sp. with sugarcane molasses as the carbon source. Food Technol. Biotechnol. (2008) 46:73–9.
181. Vamvakaki AN, Kandarakis I, Kaminarides S, Komaitis M, Papanikolaou S. Cheese whey as a renewable substrate for microbial lipid and biomass production by Zygomycetes. Eng Life Sci. (2010) 10:348–60. doi: 10.1002/elsc.201000063
182. Chan LG, Cohen JL, Ozturk G, Hennebelle M, Taha AY, Ln de Moura Bell JM. Bioconversion of cheese whey permeate into fungal oil by Mucor circinelloides. J Biol Eng. (2018) 12:25. doi: 10.1186/s13036-018-0116-5
183. Zikou E, Chatzifragkou A, Koutinas A, Papanikolaou S. Evaluating glucose and xylose as cosubstrates for lipid accumulation and γ−linolenic acid biosynthesis of Thamnidium elegans. J Appl Microbiol. (2013) 114:1020–32. doi: 10.1111/jam.12116
184. Gupta A, Abraham RE, Barrow CJ, Puri M. Omega-3 fatty acid production from enzyme saccharified hemp hydrolysate using a novel marine thraustochytrid strain. Bioresour Technol. (2015) 184:373–8. doi: 10.1016/j.biortech.2014.11.031
185. Patel A, Matsakas L, Hrůzová K, Rova U, Christakopoulos P. Biosynthesis of nutraceutical fatty acids by the oleaginous marine microalgae Phaeodactylum tricornutum utilizing hydrolysates from organosolv-pretreated birch and spruce biomass. Mar Drugs. (2019) 17:119. doi: 10.3390/md17020119
186. Iwasaka H, Aki T, Adachi H, Watanabe K, Kawamoto S, Ono K. Utilization of waste syrup for production of polyunsaturated fatty acids and xanthophylls by Aurantiochytrium. J Oleo Sci. (2013) 62:729–36. doi: 10.5650/jos.62.729
187. Yin F-W, Zhu S-Y, Guo D-S, Ren L-J, Ji X-J, Huang H, et al. Development of a strategy for the production of docosahexaenoic acid by Schizochytrium sp. from cane molasses and algae-residue. Bioresour Technol. (2019) 271:118–24. doi: 10.1016/j.biortech.2018.09.114
188. Unagul P, Assantachai C, Phadungruengluij S, Suphantharika M, Tanticharoen M, Verduyn C. Coconut water as a medium additive for the production of docosahexaenoic acid (C22: 6 n3) by Schizochytrium mangrovei Sk-02. Bioresour Technol. (2007) 98:281–7. doi: 10.1016/j.biortech.2006.01.013
189. Park W-K, Moon M, Shin S-E, Cho JM, Suh WI, Chang YK, et al. Economical DHA (Docosahexaenoic acid) production from Aurantiochytrium sp. KRS101 using orange peel extract and low cost nitrogen sources. Algal Res. (2018) 29:71–9. doi: 10.1016/j.algal.2017.11.017
190. Hong WK, Rairakhwada D, Seo PS, Park S-Y, Hur BK, Kim CH, et al. Production of lipids containing high levels of docosahexaenoic acid by a newly isolated microalga, Aurantiochytrium sp. KRS101. Appl Biochem Biotechnol. (2011) 164:1468–80. doi: 10.1007/s12010-011-9227-x
191. Silva D, Villarroel MP, Roa AL, Quilodrán BH. Use of waste from agroindustrial sources as substrate for polyunsaturated fatty acids production by Thraustochytrium Kinney VAL-B1. Int J Eng Res Afr. (2017) 33:50–5. doi: 10.4028/www.scientific.net/JERA.33.50
192. Ethier S, Woisard K, Vaughan D, Wen Z. Continuous culture of the microalgae Schizochytrium limacinum on biodiesel-derived crude glycerol for producing docosahexaenoic acid. Bioresour Technol. (2011) 102:88–93. doi: 10.1016/j.biortech.2010.05.021
193. Fakas S, Bellou S, Makri A, Aggelis G. Single cell oil and gamma-linolenic acid production by Thamnidium elegans grown on raw glycerol. In: G Aggelis editor. Microbial Conversions of Raw Glycerol. New York, NY: Nova Science Publishers (2009). p. 85–99.
194. Bellou S, Moustogianni A, Makri A, Aggelis G. Lipids containing polyunsaturated fatty acids synthesized by Zygomycetes grown on glycerol. Appl Biochem Biotechnol. (2012) 166:146–58. doi: 10.1007/s12010-011-9411-z
195. Moustogianni A, Bellou S, Triantaphyllidou IE, Aggelis G. Feasibility of raw glycerol conversion into single cell oil by zygomycetes under non-aseptic conditions. Biotechnol. Bioeng. (2015) 112:827–31. doi: 10.1002/bit.25482
196. Nicol R, Marchand K, Lubitz W. Bioconversion of crude glycerol by fungi. Appl Microbiol Biotechnol. (2012) 93:1865–75. doi: 10.1007/s00253-012-3921-7
197. Papanikolaou S, Rontou M, Belka A, Athenaki M, Gardeli C, Mallouchos A, et al. Conversion of biodiesel-derived glycerol into biotechnological products of industrial significance by yeast and fungal strains. Eng Life Sci. (2017) 17:262–81. doi: 10.1002/elsc.201500191
198. Hou CT. Production of arachidonic acid and dihomo-γ-linolenic acid from glycerol by oil-producing filamentous fungi, Mortierella in the ARS culture collection. J Ind Microbiol. (2008) 35:501–6. doi: 10.1007/s10295-008-0308-y
199. Dourou M, Tsolcha ON, Tekerlekopoulou AG, Bokas D, Aggelis G. Fish farm effluents are suitable growth media for Nannochloropsis gaditana, a polyunsaturated fatty acid producing microalga. Eng Life Sci. (2018) 18:851–60. doi: 10.1002/elsc.201800064
200. Malibari R, Sayegh F, Elazzazy AM, Baeshen MN, Dourou M, Aggelis G. Reuse of shrimp farm wastewater as growth medium for marine microalgae isolated from Red Sea–Jeddah. J Clean Prod. (2018) 198:160–9. doi: 10.1016/j.jclepro.2018.07.037
201. Brzezinska MS, Lalke-Porczyk E, Donderski W, Walczak M. Occurrence and activity of microorganisms in shrimp waste. Curr Microbiol. (2008) 57:580–7. doi: 10.1007/s00284-008-9246-1
202. Brzezinska MS, Jankiewicz U, Burkowska A, Walczak M. Chitinolytic microorganisms and their possible application in environmental protection. Curr Microbiol. (2014) 68:71–81. doi: 10.1007/s00284-013-0440-4
203. Tauk-Tornisielo SM, Arasato LS, Almeida AFd, Govone JS, Malagutti EN. Lipid formation and γ-linolenic acid production by Mucor circinelloides and Rhizopus sp., grown on vegetable oil.. Braz J Microbiol. (2009) 40:342–5. doi: 10.1590/S1517-83822009000200025
204. Sun C, Shah AM, Yang J, Wang Z, Zhu L, Song Y. Transcriptome analysis of oleaginous fungus Mucor circinelloides WJ11 in response to exogenous soybean oil as carbon source. Nat Prod Commun. (2021) 16:1934578X211023366. doi: 10.1177/1934578X211023366
205. De Souza PM, Andrade Silva NR, Souza DG, Lima e Silva TA, Freitas-Silva MC, Andrade RF. Production of a biosurfactant by Cunninghamella echinulata using renewable substrates and its applications in enhanced oil spill recovery. Colloids Interface Sci. (2018) 2:63. doi: 10.3390/colloids2040063
206. Vadivelan G, Rao PP, Venkateswaran G. Influence of supplementation of vegetable oil blends on omega-3 Fatty acid production in mortierella alpina CFR-GV15. Biomed Res Int. (2017) 2017:1432970. doi: 10.1155/2017/1432970
207. Liu X, Ashforth E, Ren B, Song F, Dai H, Liu M, et al. Bioprospecting microbial natural product libraries from the marine environment for drug discovery. J Antibiot. (2010) 63:415–22. doi: 10.1038/ja.2010.56
208. Bianchi AC, Olazábal L, Torre A, Loperena LJW. Antarctic microorganisms as source of the omega-3 polyunsaturated fatty acids. World J Microbiol Biotechnol. (2014) 30:1869–78. doi: 10.1007/s11274-014-1607-2
209. De Carvalho CC, Caramujo M. Lipids of prokaryotic origin at the base of marine food webs. Mar Drugs. (2012) 10:2698–714. doi: 10.3390/md10122698
210. Yoshida K, Hashimoto M, Hori R, Adachi T, Okuyama H, Orikasa Y, et al. Bacterial long-chain polyunsaturated fatty acids: their biosynthetic genes, functions, and practical use. Mar Drugs. (2016) 14:94. doi: 10.3390/md14050094
211. Yano Y, Nakayama A, Yoshida K. Distribution of polyunsaturated fatty acids in bacteria present in intestines of deep-sea fish and shallow-sea poikilothermic animals. Appl Environ Microbiol. (1997) 63:2572–7. doi: 10.1128/aem.63.7.2572-2577.1997
212. Gupta A, Barrow CJ, Puri M. Omega-3 biotechnology: thraustochytrids as a novel source of omega-3 oils. Biotechnol Adv. (2012) 30:1733–45. doi: 10.1016/j.biotechadv.2012.02.014
213. Das S, Lyla P, Khan SA. Marine microbial diversity and ecology: importance and future perspectives. Curr Sci. (2006) 90:1325–35.
214. Gao W, Kim JY, Anderson JR, Akopian T, Hong S, Jin Y-Y, et al. The cyclic peptide ecumicin targeting ClpC1 is active against Mycobacterium tuberculosis in vivo. Antimicrob Agents Chemother. (2015) 59:880–9. doi: 10.1128/AAC.04054-14
215. Wang F, Wang P, Chen M, Xiao X. Isolation of extremophiles with the detection and retrieval of Shewanella strains in deep-sea sediments from the west Pacific. Extremophiles. (2004) 8:165–8. doi: 10.1007/s00792-003-0365-0
216. Usui K, Hiraki T, Kawamoto J, Kurihara T, Nogi Y, Kato C, et al. Eicosapentaenoic acid plays a role in stabilizing dynamic membrane structure in the deep-sea piezophile Shewanella violacea: a study employing high-pressure time-resolved fluorescence anisotropy measurement. Biochim Biophys Acta. (2012) 1818:574–83. doi: 10.1016/j.bbamem.2011.10.010
217. Nogi Y, Kato C, Horikoshi K. Taxonomic studies of deep-sea barophilic Shewanella strains and description of Shewanella violacea sp. nov. Arch Microbiol. (1998) 170:331–8. doi: 10.1007/s002030050650
218. Estupiñán M, Hernández I, Saitua E, Bilbao ME, Mendibil I, Ferrer J, et al. Novel Vibrio spp. strains producing omega-3 fatty acids isolated from coastal seawater. Mar Drugs. (2020) 18:99. doi: 10.3390/md18020099
219. Yang HL, Lu C-K, Chen SF, Chen YM, Chen YM. Isolation and characterization of Taiwanese heterotrophic microalgae: screening of strains for docosahexaenoic acid (DHA) production. Mar Biotechnol. (2010) 12:173–85. doi: 10.1007/s10126-009-9207-0
220. Kimura H, Fukuba T, Naganuma T. Biomass of thraustochytrid protoctists in coastal water. Mar Ecol Prog Ser. (1999) 189:27–33. doi: 10.3354/meps189027
221. Burja AM, Radianingtyas H, Windust A, Barrow C. Isolation and characterization of polyunsaturated fatty acid producing Thraustochytrium species: screening of strains and optimization of omega-3 production. Appl Microbiol Biotechnol. (2006) 72:1161–9. doi: 10.1007/s00253-006-0419-1
222. Raghukumar S. Ecology of the marine protists, the Labyrinthulomycetes (Thraustochytrids and Labyrinthulids). Eur J Protistol. (2002) 38:127–45. doi: 10.1078/0932-4739-00832
223. Schloss PD, Handelsman J. Status of the microbial census. Microbiol Mol Biol Rev. (2004) 68:686–91. doi: 10.1128/MMBR.68.4.686-691.2004
224. Rappé MS, Giovannoni S. The uncultured microbial majority. Uncult Microb Major. (2003) 57:369–94. doi: 10.1146/annurev.micro.57.030502.090759
225. Pathom-Aree W, Stach JE, Ward AC, Horikoshi K, Bull AT, Goodfellow M. Diversity of actinomycetes isolated from Challenger Deep sediment (10,898 m) from the Mariana Trench. Extremophiles. (2006) 10:181–9. doi: 10.1007/s00792-005-0482-z
226. Knight V, Sanglier J-J, DiTullio D, Braccili S, Bonner P, Waters J, et al. Diversifying microbial natural products for drug discovery. Appl Microbiol Biotechnol. (2003) 62:446–58. doi: 10.1007/s00253-003-1381-9
227. Koehn FE. High impact technologies for natural products screening. Nat Compou Drugs Vol. (2008) 65:175–210. doi: 10.1007/978-3-7643-8117-2_5
228. Miranda C, Bettencourt S, Pozdniakova T, Pereira J, Sampaio P, Franco-Duarte R, et al. Modified high-throughput Nile red fluorescence assay for the rapid screening of oleaginous yeasts using acetic acid as carbon source. BMC Microbiol. (2020) 20:60. doi: 10.1186/s12866-020-01742-6
229. Wang L, Chen W, Feng Y, Ren Y, Gu Z, Chen H, et al. Genome characterization of the oleaginous fungus Mortierella alpina. PLoS One. (2011) 6:e28319. doi: 10.1371/journal.pone.0028319
230. Rossi M, Buzzini P, Cordisco L, Amaretti A, Sala M, Raimondi S, et al. Growth, lipid accumulation, and fatty acid composition in obligate psychrophilic, facultative psychrophilic, and mesophilic yeasts. FEMS Microbiol. Ecol. (2009) 69:363–72. doi: 10.1111/j.1574-6941.2009.00727.x
231. Ageitos JM, Vallejo JA, Veiga-Crespo P, Villa TG. Oily yeasts as oleaginous cell factories. Appl Microbiol Biotechnol. (2011) 90:1219–27. doi: 10.1007/s00253-011-3200-z
232. Rumin J, Bonnefond H, Saint-Jean B, Rouxel C, Sciandra A, Bernard O, et al. The use of fluorescent Nile red and BODIPY for lipid measurement in microalgae. Biotechnol Biofuels. (2015) 8:1–16. doi: 10.1186/s13068-015-0220-4
233. Bligh EG, Dyer W. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. (1959) 37:911–7. doi: 10.1139/o59-099
234. Cooksey KE, Guckert JB, Williams SA, Callis PR. Fluorometric determination of the neutral lipid content of microalgal cells using Nile Red. J Microbiol Methods. (1987) 6:333–45. doi: 10.1016/0167-7012(87)90019-4
235. Sitepu I, Ignatia L, Franz A, Wong D, Faulina S, Tsui M, et al. An improved high-throughput Nile red fluorescence assay for estimating intracellular lipids in a variety of yeast species. J Microbiol Methods. (2012) 91:321–8. doi: 10.1016/j.mimet.2012.09.001
236. Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol. (1985) 100:965–73. doi: 10.1083/jcb.100.3.965
237. Ami D, Posteri R, Mereghetti P, Porro D, Doglia SM, Branduardi P. Fourier transform infrared spectroscopy as a method to study lipid accumulation in oleaginous yeasts. Biotechnol Biofuels. (2014) 7:12. doi: 10.1186/1754-6834-7-12
238. Rostron KA, Rolph CE, Lawrence CL. Nile red fluorescence screening facilitating neutral lipid phenotype determination in budding yeast, Saccharomyces cerevisiae, and the fission yeast Schizosaccharomyces pombe. Antonie Van Leeuwenhoek. (2015) 108:97–106. doi: 10.1007/s10482-015-0467-6
239. O’Reilly A, Scott J. Defined coimmobilization of mixed microorganism cultures. Enzyme Microb Technol. (1995) 17:636–46. doi: 10.1016/0141-0229(94)00103-X
240. Ji M-K, Kim HC, Sapireddy VR, Yun HS, Abou-Shanab RA, Choi J, et al. Simultaneous nutrient removal and lipid production from pretreated piggery wastewater by Chlorella vulgaris YSW-04. Appl Microbiol Biotechnol. (2013) 97:2701–10. doi: 10.1007/s00253-012-4097-x
241. Hongyang S, Yalei Z, Chunmin Z, Xuefei Z, Jinpeng L. Cultivation of Chlorella pyrenoidosa in soybean processing wastewater. Bioresour Technol. (2011) 102:9884–90. doi: 10.1016/j.biortech.2011.08.016
242. Müller R, Wink J. Future potential for anti-infectives from bacteria–how to exploit biodiversity and genomic potential. Int J Med Microbiol. (2014) 304:3–13. doi: 10.1016/j.ijmm.2013.09.004
243. Chiang YM, Chang SL, Oakley BR, Wang CC. Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr Opin Chem Biol. (2011) 15:137–43. doi: 10.1016/j.cbpa.2010.10.011
244. De-Bashan LE, Bashan Y, Moreno M, Lebsky VK, Bustillos J. Increased pigment and lipid content, lipid variety, and cell and population size of the microalgae Chlorella spp. when co-immobilized in alginate beads with the microalgae-growth-promoting bacterium Azospirillum brasilense. Can J Microbiol. (2002) 48:514–21. doi: 10.1139/w02-051
245. Cai S, Hu C, Du S. Comparisons of growth and biochemical composition between mixed culture of alga and yeast and monocultures. J Biosci Bioeng. (2007) 104:391–7. doi: 10.1263/jbb.104.391
246. Xue F, Miao J, Zhang X, Tan T. A new strategy for lipid production by mix cultivation of Spirulina platensis and Rhodotorula glutinis. Appl Biochem Biotechnol. (2010) 160:498–503. doi: 10.1007/s12010-008-8376-z
247. Cheirsilp B, Kitcha S, Torpee SJ. Co-culture of an oleaginous yeast Rhodotorula glutinis and a microalga Chlorella vulgaris for biomass and lipid production using pure and crude glycerol as a sole carbon source. Ann Microbiol. (2012) 62:987–93. doi: 10.1007/s13213-011-0338-y
248. Zhao P, Yu X, Li J, Tang X, Huang Z. Enhancing lipid productivity by co-cultivation of chlorella sp. U4341 and Monoraphidium sp. FXY-10. J Biosci Bioeng. (2014) 118:72–7. doi: 10.1016/j.jbiosc.2013.12.014
249. Cheirsilp B, Suwannarat W, Niyomdecha R. Mixed culture of oleaginous yeast Rhodotorula glutinis and microalga Chlorella vulgaris for lipid production from industrial wastes and its use as biodiesel feedstock. N Biotechnol. (2011) 28:362–8. doi: 10.1016/j.nbt.2011.01.004
250. Dostálek M, Häggström MH. Mixed culture of Saccharomycopsis fibuliger and Zymomonas mobilis on starch-use of oxygen as a regulator. Eur J Appl Microbiol Biotechnol. (1983) 17:269–74. doi: 10.1007/BF00508019
251. Abdelmohsen UR, Grkovic T, Balasubramanian S, Kamel MS, Quinn RJ, Hentschel U. Elicitation of secondary metabolism in actinomycetes. Biotechnol Adv. (2015) 33:798–811. doi: 10.1016/j.biotechadv.2015.06.003
252. Katsuyama Y. Mining novel biosynthetic machineries of secondary metabolites from actinobacteria. Biosci Biotechnol Biochem. (2019) 83:1606–15. doi: 10.1080/09168451.2019.1606700
253. Zhu X, Li S, Liu L, Li S, Luo Y, Lv C, et al. Genome sequencing and analysis of Thraustochytriidae sp. SZU445 provides novel insights into the polyunsaturated fatty acid biosynthesis pathway. Mar Drugs. (2020) 18:118. doi: 10.3390/md18020118
254. Song Z, Stajich JE, Xie Y, Liu X, He Y, Chen J, et al. Comparative analysis reveals unexpected genome features of newly isolated Thraustochytrids strains: on ecological function and PUFAs biosynthesis. BMC Genom. (2018) 19:541. doi: 10.1186/s12864-018-4904-6
255. Schneider O, Ilic-Tomic T, Rückert C, Kalinowski J, Genčić MS, Živković MZ, et al. Genomics-based insights into the biosynthesis and unusually high accumulation of free fatty acids by Streptomyces sp. NP10. Front Microbiol. (2018) 9:1302. doi: 10.3389/fmicb.2018.01302
256. Müller R. Don’t classify polyketide synthases. Chem Biol. (2004) 11:4–6. doi: 10.1016/j.chembiol.2004.01.005
257. Albataineh H, Stevens DC. Marine myxobacteria: a few good halophiles. Mar Drugs. (2018) 16:209. doi: 10.3390/md16060209
258. Reichenbach H. Order VIII. myxococcales tchan, portion and prevot 1948, 398. Bergeys Manu Syst Bacteriol. (2005) 2:1059–72.
259. Gemperlein K, Rachid S, Garcia RO, Wenzel SC, Müller R. Polyunsaturated fatty acid biosynthesis in myxobacteria: different PUFA synthases and their product diversity. Chem Sci. (2014) 5:1733–41. doi: 10.1039/c3sc53163e
260. Garcia R, Pistorius D, Stadler M, Müller R. Fatty acid-related phylogeny of myxobacteria as an approach to discover polyunsaturated omega-3/6 fatty acids. J Bacteriol. (2011) 193:1930–42. doi: 10.1128/JB.01091-10
261. Iizuka T, Jojima Y, Fudou R, Tokura M, Hiraishi A, Yamanaka S. Enhygromyxa salina gen. nov., sp. nov., a slightly halophilic myxobacterium isolated from the coastal areas of Japan. Syst Appl Microbiol. (2003) 26:189–96. doi: 10.1078/072320203322346038
262. Amiri Moghaddam J, Poehlein A, Fisch K, Alanjary M, Daniel R, König GM, et al. Draft genome sequences of the obligatory marine myxobacterial strains Enhygromyxa salina SWB005 and SWB007. Genome Announc. (2018) 6:e324–318. doi: 10.1128/genomeA.00324-18
263. Ando A, Sumida Y, Negoro H, Suroto DA, Ogawa J, Sakuradani E, et al. Establishment of Agrobacterium tumefaciens-mediated transformation of an oleaginous fungus, Mortierella alpina 1S-4, and its application for eicosapentaenoic acid producer breeding. Appl Environ Microbiol. (2009) 75:5529–35. doi: 10.1128/AEM.00648-09
264. Zhao G, Yao Y, Chen W, Cao X. Comparison and analysis of the genomes of two Aspergillus oryzae strains. J Agric Food Chem. (2013) 61:7805–9. doi: 10.1021/jf400080g
265. Tang X, Zhao L, Chen H, Chen YQ, Chen W, Song Y, et al. Complete genome sequence of a high lipid-producing strain of Mucor circinelloides WJ11 and comparative genome analysis with a low lipid-producing strain CBS 277.49. PLoS One. (2015) 10:e0137543. doi: 10.1371/journal.pone.0137543
266. Ratledge C. Microbial production of polyunsaturated fatty acids as nutraceuticals. In: B McNeil, D Archer, I Giavasis, L Harvey editors. Microbial Production of Food Ingredients, Enzymes and Nutraceuticals. Cambridge: Woodhead Publishing (2013). p. 531–58. doi: 10.1533/9780857093547.2.531
267. Sawangkeaw R, Ngamprasertsith S. A review of lipid-based biomasses as feedstocks for biofuels production, renew. Sust Energy Rev. (2013) 25:97–108. doi: 10.1016/j.rser.2013.04.007
268. Thevenieau F, Nicaud JM. Microorganisms as sources of oils. OCL. (2013) 20:D603. doi: 10.1051/ocl/2013034
269. Abeln F, Chuck CJ. The history, state of the art and future prospects for oleaginous yeast research. Microb Cell Fact. (2021) 20:1–31. doi: 10.1186/s12934-021-01712-1
270. Karamerou EE, Parsons S, McManus MC, Chuck CJ. Using techno-economic modelling to determine the minimum cost possible for a microbial palm oil substitute. Biotechnol Biofuels. (2021) 14:1–9. doi: 10.1186/s13068-021-01911-3
Keywords: polyunsaturated fatty acids, novel oleaginous microbes, metabolic engineering, genome mining, agro - industrial waste conversion
Citation: Shah AM, Yang W, Mohamed H, Zhang Y and Song Y (2022) Microbes: A Hidden Treasure of Polyunsaturated Fatty Acids. Front. Nutr. 9:827837. doi: 10.3389/fnut.2022.827837
Received: 22 December 2021; Accepted: 21 February 2022;
Published: 17 March 2022.
Edited by:
Hani El-Nezami, The University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Jetty Chung-Yung Lee, The University of Hong Kong, Hong Kong SAR, ChinaXiao-Jun Ji, Nanjing Tech University, China
Seraphim Papanikolaou, Agricultural University of Athens, Greece
Copyright © 2022 Shah, Yang, Mohamed, Zhang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingtong Zhang, ytzhang@jaas.ac.cn; Yuanda Song, ysong@sdut.edu.cn
†These authors have contributed equally to this work and share first authorship
 Aabid Manzoor Shah1†
Aabid Manzoor Shah1†  Hassan Mohamed
Hassan Mohamed Yingtong Zhang
Yingtong Zhang Yuanda Song
Yuanda Song